UniProtKB/Swiss-Prot protein knowledgebase release 2012_10 statistics
1. INTRODUCTION
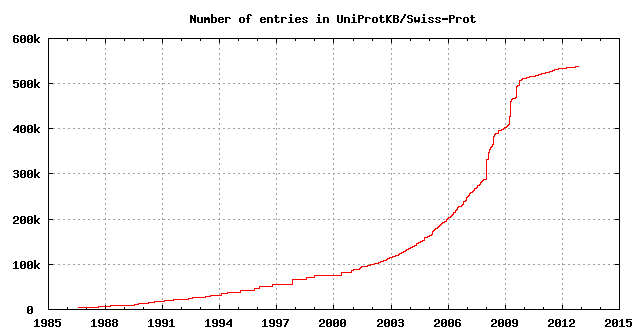
Release 2012_10 of 31-Oct-12 of UniProtKB/Swiss-Prot contains 538259 sequence entries,
comprising 191113170 amino acids abstracted from 214310 references.
251 sequences have been added since release 2012_09, the sequence data of
94 existing entries has been updated and the annotations of
101771 entries have been revised.
Number of fragments: 9040
Number of additional sequences produced by alternative splicing, initiation or promoter usage, or ribosomal frameshifting: 32585
Protein existence (PE): entries %
1: Evidence at protein level 76560 14.2%
2: Evidence at transcript level 68097 12.7%
3: Inferred from homology 377381 70.1%
4: Predicted 14332 2.7%
5: Uncertain 1889 0.4%
The growth of the database is summarized below.
 2. TAXONOMIC ORIGIN
Total number of species represented in this release of UniProtKB/Swiss-Prot: 12922
The first twenty species represent 112553 sequences: 20.9 % of the total
number of entries.
2.1 Table of the frequency of occurrence of species
Species represented 1x: 5426
2x: 1882
3x: 981
4x: 639
5x: 466
6x: 381
7x: 284
8x: 217
9x: 199
10x: 123
11- 20x: 668
21- 50x: 403
51-100x: 212
>100x: 1041
2.2 Table of the most represented species
------ --------- --------------------------------------------
Number Frequency Species
------ --------- --------------------------------------------
1 20233 Homo sapiens (Human)
2 16566 Mus musculus (Mouse)
3 11571 Arabidopsis thaliana (Mouse-ear cress)
4 7815 Rattus norvegicus (Rat)
5 6621 Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
6 5965 Bos taurus (Bovine)
7 5089 Schizosaccharomyces pombe (strain 972 / ATCC 24843) (Fission yeast)
8 4431 Escherichia coli (strain K12)
9 4188 Bacillus subtilis (strain 168)
10 4126 Dictyostelium discoideum (Slime mold)
11 3364 Caenorhabditis elegans
12 3356 Xenopus laevis (African clawed frog)
13 3165 Drosophila melanogaster (Fruit fly)
14 2967 Oryza sativa subsp. japonica (Rice)
15 2861 Danio rerio (Zebrafish) (Brachydanio rerio)
16 2252 Gallus gallus (Chicken)
17 2218 Pongo abelii (Sumatran orangutan) (Pongo pygmaeus abelii)
18 2012 Escherichia coli O157:H7
19 1966 Mycobacterium tuberculosis
20 1787 Methanocaldococcus jannaschii
21 1770 Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)
22 1707 Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd)
23 1678 Shigella flexneri
24 1678 Xenopus tropicalis (Western clawed frog) (Silurana tropicalis)
25 1675 Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC)
26 1409 Sus scrofa (Pig)
27 1346 Salmonella typhi
28 1242 Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 / 1C / PRS 101 / LMG 12228)
29 1241 Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97)
30 1170 Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
31 1036 Synechocystis sp. (strain PCC 6803 / Kazusa)
32 1018 Yersinia pestis
33 1012 Archaeoglobus fulgidus
34 949 Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)
35 930 Salmonella paratyphi A (strain ATCC 9150 / SARB42)
36 926 Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056)
37 925 Staphylococcus aureus (strain N315)
38 923 Staphylococcus aureus (strain Mu50 / ATCC 700699)
39 909 Acanthamoeba polyphaga mimivirus (APMV)
40 904 Kluyveromyces lactis
41 899 Staphylococcus aureus (strain COL)
42 895 Staphylococcus aureus (strain MW2)
43 889 Staphylococcus aureus (strain MSSA476)
44 888 Escherichia coli O6:K15:H31 (strain 536 / UPEC)
45 888 Staphylococcus aureus (strain MRSA252)
46 886 Oryctolagus cuniculus (Rabbit)
47 882 Salmonella choleraesuis (strain SC-B67)
48 878 Shigella sonnei (strain Ss046)
49 869 Rhizobium meliloti (strain 1021) (Ensifer meliloti) (Sinorhizobium meliloti)
50 863 Yersinia pseudotuberculosis serotype I (strain IP32953)
51 861 Candida glabrata
52 841 Escherichia coli O9:H4 (strain HS)
53 834 Neurospora crassa
54 834 Escherichia coli O139:H28 (strain E24377A / ETEC)
55 829 Shigella boydii serotype 4 (strain Sb227)
56 825 Escherichia coli (strain UTI89 / UPEC)
57 821 Shigella dysenteriae serotype 1 (strain Sd197)
58 819 Escherichia coli (strain ATCC 8739 / DSM 1576 / Crooks)
59 803 Canis familiaris (Dog) (Canis lupus familiaris)
60 791 Escherichia coli (strain SMS-3-5 / SECEC)
61 787 Vibrio parahaemolyticus serotype O3:K6 (strain RIMD 2210633)
62 783 Erwinia carotovora subsp. atroseptica (strain SCRI 1043 / ATCC BAA-672)
63 782 Aquifex aeolicus (strain VF5)
64 775 Pasteurella multocida (strain Pm70)
65 773 Emericella nidulans
66 771 Escherichia coli (strain K12 / DH10B)
67 765 Escherichia coli O127:H6 (strain E2348/69 / EPEC)
68 765 Escherichia coli (strain K12 / MC4100 / BW2952)
69 764 Escherichia coli O17:K52:H18 (strain UMN026 / ExPEC)
70 762 Escherichia coli (strain 55989 / EAEC)
71 761 Escherichia coli O8 (strain IAI1)
72 760 Shigella flexneri serotype 5b (strain 8401)
73 759 Staphylococcus epidermidis (strain ATCC 35984 / RP62A)
74 758 Staphylococcus epidermidis (strain ATCC 12228)
75 756 Escherichia coli (strain SE11)
76 756 Escherichia coli O45:K1 (strain S88 / ExPEC)
77 754 Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145)
78 753 Escherichia coli O7:K1 (strain IAI39 / ExPEC)
79 748 Escherichia coli O157:H7 (strain EC4115 / EHEC)
80 744 Photorhabdus luminescens subsp. laumondii (strain TT01)
81 740 Staphylococcus aureus (strain NCTC 8325)
82 735 Bacillus halodurans
83 735 Yersinia enterocolitica serotype O:8 / biotype 1B (strain 8081)
84 734 Bacillus anthracis
85 732 Vibrio vulnificus (strain CMCP6)
86 731 Escherichia coli O81 (strain ED1a)
87 721 Salmonella enteritidis PT4 (strain P125109)
88 717 Vibrio vulnificus (strain YJ016)
89 716 Salmonella paratyphi B (strain ATCC BAA-1250 / SPB7)
90 715 Yersinia pestis bv. Antiqua (strain Nepal516)
91 714 Salmonella paratyphi A (strain AKU_12601)
92 713 Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578)
93 713 Enterobacter sp. (strain 638)
94 713 Salmonella agona (strain SL483)
95 713 Escherichia coli O1:K1 / APEC
96 713 Salmonella newport (strain SL254)
97 713 Yersinia pseudotuberculosis serotype O:1b (strain IP 31758)
98 712 Salmonella schwarzengrund (strain CVM19633)
99 711 Yersinia pestis bv. Antiqua (strain Antiqua)
100 711 Zea mays (Maize)
101 710 Salmonella heidelberg (strain SL476)
102 702 Salmonella dublin (strain CT_02021853)
103 701 Candida albicans (strain SC5314 / ATCC MYA-2876) (Yeast)
104 698 Shigella boydii serotype 18 (strain CDC 3083-94 / BS512)
105 696 Klebsiella pneumoniae (strain 342)
106 695 Escherichia fergusonii (strain ATCC 35469 / DSM 13698 / CDC 0568-73)
107 693 Pseudomonas putida (strain KT2440)
108 689 Nostoc sp. (strain PCC 7120 / UTEX 2576)
109 688 Pan troglodytes (Chimpanzee)
110 687 Mycoplasma pneumoniae (strain ATCC 29342 / M129)
111 683 Salmonella gallinarum (strain 287/91 / NCTC 13346)
112 678 Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
113 675 Pseudomonas syringae pv. tomato (strain DC3000)
114 670 Serratia proteamaculans (strain 568)
115 668 Mycobacterium leprae (strain TN)
116 667 Yersinia pestis (strain Pestoides F)
117 666 Staphylococcus aureus (strain USA300)
118 658 Rhizobium sp. (strain NGR234)
119 654 Bradyrhizobium japonicum (strain USDA 110)
120 653 Debaryomyces hansenii
121 652 Bacillus cereus (strain ATCC 14579 / DSM 31)
122 650 Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100)
123 645 Escherichia coli
124 643 Staphylococcus aureus (strain bovine RF122 / ET3-1)
125 642 Salmonella arizonae (strain ATCC BAA-731 / CDC346-86 / RSK2980)
126 642 Yarrowia lipolytica (strain CLIB 122 / E 150) (Yeast) (Candida lipolytica)
127 638 Yersinia pseudotuberculosis serotype O:3 (strain YPIII)
128 634 Shewanella oneidensis (strain MR-1)
129 634 Yersinia pseudotuberculosis serotype IB (strain PB1/+)
130 632 Agrobacterium tumefaciens (strain C58 / ATCC 33970)
131 622 Cronobacter sakazakii (strain ATCC BAA-894) (Enterobacter sakazakii)
132 616 Treponema pallidum (strain Nichols)
133 615 Oryza sativa subsp. indica (Rice)
134 613 Methanothermobacter thermautotrophicus
135 612 Staphylococcus haemolyticus (strain JCSC1435)
136 606 Rhizobium loti (strain MAFF303099) (Mesorhizobium loti)
137 605 Xanthomonas campestris pv. campestris (strain ATCC 33913 / NCPPB 528 / LMG 568)
138 602 Ralstonia solanacearum (strain GMI1000) (Pseudomonas solanacearum)
139 602 Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e)
140 602 Photobacterium profundum (Photobacterium sp. (strain SS9))
141 602 Staphylococcus saprophyticus subsp. saprophyticus
142 601 Salmonella paratyphi C (strain RKS4594)
143 600 Yersinia pestis bv. Antiqua (strain Angola)
144 590 Bacillus cereus (strain ATCC 10987)
145 590 Listeria innocua serovar 6a (strain CLIP 11262)
146 589 Pectobacterium carotovorum subsp. carotovorum (strain PC1)
147 586 Rickettsia prowazekii (strain Madrid E)
148 580 Helicobacter pylori (strain ATCC 700392 / 26695) (Campylobacter pylori)
149 578 Neisseria meningitidis serogroup B (strain MC58)
150 576 Brucella suis biovar 1 (strain 1330)
151 572 Brucella melitensis biotype 1 (strain 16M / ATCC 23456 / NCTC 10094)
152 572 Buchnera aphidicola subsp. Acyrthosiphon pisum (strain APS)
153 568 Caenorhabditis briggsae
154 567 Bacillus thuringiensis subsp. konkukian (strain 97-27)
155 566 Pseudomonas syringae pv. syringae (strain B728a)
156 565 Helicobacter pylori (strain J99) (Campylobacter pylori J99)
157 564 Vibrio fischeri (strain ATCC 700601 / ES114)
158 564 Caulobacter crescentus (strain ATCC 19089 / CB15)
159 564 Pseudomonas aeruginosa (strain UCBPP-PA14)
160 562 Bacillus licheniformis (strain DSM 13 / ATCC 14580)
161 562 Buchnera aphidicola subsp. Schizaphis graminum (strain Sg)
162 561 Bacillus cereus (strain ZK / E33L)
163 556 Xanthomonas axonopodis pv. citri (strain 306)
164 556 Clostridium acetobutylicum
165 552 Oceanobacillus iheyensis (strain DSM 14371 / JCM 11309 / KCTC 3954 / HTE831)
166 552 Neisseria meningitidis serogroup A / serotype 4A (strain Z2491)
167 552 Pseudomonas fluorescens (strain Pf0-1)
168 546 Pseudomonas fluorescens (strain Pf-5 / ATCC BAA-477)
169 545 Pseudomonas syringae pv. phaseolicola (strain 1448A / Race 6)
170 533 Lactococcus lactis subsp. lactis (strain IL1403) (Streptococcus lactis)
171 531 Erwinia tasmaniensis (strain DSM 17950 / Et1/99)
172 529 Sodalis glossinidius (strain morsitans)
173 529 Listeria monocytogenes serotype 4b (strain F2365)
174 527 Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099)
175 522 Bordetella bronchiseptica (strain ATCC BAA-588 / NCTC 13252 / RB50)
176 522 Xylella fastidiosa (strain 9a5c)
177 515 Chromobacterium violaceum
178 515 Bordetella pertussis (strain Tohama I / ATCC BAA-589 / NCTC 13251)
179 512 Xylella fastidiosa (strain Temecula1 / ATCC 700964)
180 511 Pseudomonas aeruginosa (strain PA7)
181 511 Vibrio cholerae serotype O1 (strain ATCC 39541 / Ogawa 395 / O395)
182 510 Haemophilus ducreyi (strain 35000HP / ATCC 700724)
183 508 Bordetella parapertussis (strain 12822 / ATCC BAA-587 / NCTC 13253)
184 507 Buchnera aphidicola subsp. Baizongia pistaciae (strain Bp)
185 507 Geobacillus kaustophilus (strain HTA426)
186 506 Staphylococcus aureus (strain Newman)
187 502 Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4)
188 501 Deinococcus radiodurans
189 500 Pseudomonas entomophila (strain L48)
190 499 Corynebacterium glutamicum
191 499 Brucella abortus biovar 1 (strain 9-941)
192 497 Streptomyces avermitilis
193 497 Rickettsia conorii (strain ATCC VR-613 / Malish 7)
194 496 Bacillus clausii (strain KSM-K16)
195 495 Burkholderia pseudomallei (strain K96243)
196 495 Haemophilus influenzae (strain 86-028NP)
197 494 Proteus mirabilis (strain HI4320)
198 492 Bacillus amyloliquefaciens (strain FZB42)
199 491 Xanthomonas campestris pv. campestris (strain 8004)
200 490 Vibrio harveyi (strain ATCC BAA-1116 / BB120)
201 489 Methanosarcina acetivorans (strain ATCC 35395 / DSM 2834 / JCM 12185 / C2A)
202 487 Shewanella sp. (strain MR-7)
203 486 Mannheimia succiniciproducens (strain MBEL55E)
204 484 Pseudomonas aeruginosa (strain LESB58)
205 484 Staphylococcus aureus (strain Mu3 / ATCC 700698)
206 484 Shewanella sp. (strain MR-4)
207 483 Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195)
208 481 Thermosynechococcus elongatus (strain BP-1)
209 480 Acinetobacter sp. (strain ADP1)
210 479 Pyrococcus horikoshii
211 478 Synechococcus elongatus (strain PCC 7942) (Anacystis nidulans R2)
212 475 Pseudomonas putida (strain F1 / ATCC 700007)
213 474 Burkholderia sp. (strain 383) (Burkholderia cepacia
214 474 Brucella abortus (strain 2308)
215 473 Aspergillus oryzae (strain ATCC 42149 / RIB 40) (Yellow koji mold)
216 472 Streptococcus pneumoniae (strain ATCC BAA-255 / R6)
217 469 Methanosarcina mazei
218 467 Halobacterium salinarum (strain ATCC 700922 / JCM 11081 / NRC-1)
219 467 Clostridium perfringens (strain 13 / Type A)
220 466 Pyrococcus abyssi (strain GE5 / Orsay)
221 466 Xanthomonas campestris pv. vesicatoria (strain 85-10)
222 466 Shewanella frigidimarina (strain NCIMB 400)
223 466 Pseudomonas putida (strain GB-1)
224 466 Cupriavidus necator (strain ATCC 17699 / H16 / DSM 428 / Stanier 337)
225 464 Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / NCIB 9240)
226 463 Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1)
227 463 Shewanella sp. (strain ANA-3)
228 462 Burkholderia mallei (strain ATCC 23344)
229 462 Anabaena variabilis (strain ATCC 29413 / PCC 7937)
230 462 Rhodopseudomonas palustris (strain ATCC BAA-98 / CGA009)
231 461 Lactobacillus plantarum (strain ATCC BAA-793 / NCIMB 8826 / WCFS1)
232 459 Cupriavidus pinatubonensis (strain JMP134 / LMG 1197) (Alcaligenes eutrophus)
233 458 Enterococcus faecalis (strain ATCC 700802 / V583)
234 455 Staphylococcus aureus (strain JH1)
235 455 Rhodobacter sphaeroides (strain ATCC 17023 / 2.4.1 / NCIB 8253 / DSM 158)
236 454 Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath)
237 454 Xanthomonas oryzae pv. oryzae (strain MAFF 311018)
238 453 Ovis aries (Sheep)
239 453 Pseudomonas putida (strain W619)
240 453 Rickettsia felis (strain ATCC VR-1525 / URRWXCal2) (Rickettsia azadi)
241 452 Shewanella baltica (strain OS185)
242 451 Aeromonas salmonicida (strain A449)
243 449 Thermoanaerobacter tengcongensis
244 449 Staphylococcus aureus (strain JH9)
245 449 Hahella chejuensis (strain KCTC 2396)
246 448 Nicotiana tabacum (Common tobacco)
247 448 Mycobacterium paratuberculosis (strain ATCC BAA-968 / K-10)
248 448 Streptococcus mutans serotype c (strain ATCC 700610 / UA159)
249 447 Vibrio fischeri (strain MJ11)
250 446 Sulfolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2)
2.3 Taxonomic distribution of the sequences
2. TAXONOMIC ORIGIN
Total number of species represented in this release of UniProtKB/Swiss-Prot: 12922
The first twenty species represent 112553 sequences: 20.9 % of the total
number of entries.
2.1 Table of the frequency of occurrence of species
Species represented 1x: 5426
2x: 1882
3x: 981
4x: 639
5x: 466
6x: 381
7x: 284
8x: 217
9x: 199
10x: 123
11- 20x: 668
21- 50x: 403
51-100x: 212
>100x: 1041
2.2 Table of the most represented species
------ --------- --------------------------------------------
Number Frequency Species
------ --------- --------------------------------------------
1 20233 Homo sapiens (Human)
2 16566 Mus musculus (Mouse)
3 11571 Arabidopsis thaliana (Mouse-ear cress)
4 7815 Rattus norvegicus (Rat)
5 6621 Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
6 5965 Bos taurus (Bovine)
7 5089 Schizosaccharomyces pombe (strain 972 / ATCC 24843) (Fission yeast)
8 4431 Escherichia coli (strain K12)
9 4188 Bacillus subtilis (strain 168)
10 4126 Dictyostelium discoideum (Slime mold)
11 3364 Caenorhabditis elegans
12 3356 Xenopus laevis (African clawed frog)
13 3165 Drosophila melanogaster (Fruit fly)
14 2967 Oryza sativa subsp. japonica (Rice)
15 2861 Danio rerio (Zebrafish) (Brachydanio rerio)
16 2252 Gallus gallus (Chicken)
17 2218 Pongo abelii (Sumatran orangutan) (Pongo pygmaeus abelii)
18 2012 Escherichia coli O157:H7
19 1966 Mycobacterium tuberculosis
20 1787 Methanocaldococcus jannaschii
21 1770 Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)
22 1707 Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd)
23 1678 Shigella flexneri
24 1678 Xenopus tropicalis (Western clawed frog) (Silurana tropicalis)
25 1675 Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC)
26 1409 Sus scrofa (Pig)
27 1346 Salmonella typhi
28 1242 Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 / 1C / PRS 101 / LMG 12228)
29 1241 Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97)
30 1170 Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
31 1036 Synechocystis sp. (strain PCC 6803 / Kazusa)
32 1018 Yersinia pestis
33 1012 Archaeoglobus fulgidus
34 949 Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)
35 930 Salmonella paratyphi A (strain ATCC 9150 / SARB42)
36 926 Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056)
37 925 Staphylococcus aureus (strain N315)
38 923 Staphylococcus aureus (strain Mu50 / ATCC 700699)
39 909 Acanthamoeba polyphaga mimivirus (APMV)
40 904 Kluyveromyces lactis
41 899 Staphylococcus aureus (strain COL)
42 895 Staphylococcus aureus (strain MW2)
43 889 Staphylococcus aureus (strain MSSA476)
44 888 Escherichia coli O6:K15:H31 (strain 536 / UPEC)
45 888 Staphylococcus aureus (strain MRSA252)
46 886 Oryctolagus cuniculus (Rabbit)
47 882 Salmonella choleraesuis (strain SC-B67)
48 878 Shigella sonnei (strain Ss046)
49 869 Rhizobium meliloti (strain 1021) (Ensifer meliloti) (Sinorhizobium meliloti)
50 863 Yersinia pseudotuberculosis serotype I (strain IP32953)
51 861 Candida glabrata
52 841 Escherichia coli O9:H4 (strain HS)
53 834 Neurospora crassa
54 834 Escherichia coli O139:H28 (strain E24377A / ETEC)
55 829 Shigella boydii serotype 4 (strain Sb227)
56 825 Escherichia coli (strain UTI89 / UPEC)
57 821 Shigella dysenteriae serotype 1 (strain Sd197)
58 819 Escherichia coli (strain ATCC 8739 / DSM 1576 / Crooks)
59 803 Canis familiaris (Dog) (Canis lupus familiaris)
60 791 Escherichia coli (strain SMS-3-5 / SECEC)
61 787 Vibrio parahaemolyticus serotype O3:K6 (strain RIMD 2210633)
62 783 Erwinia carotovora subsp. atroseptica (strain SCRI 1043 / ATCC BAA-672)
63 782 Aquifex aeolicus (strain VF5)
64 775 Pasteurella multocida (strain Pm70)
65 773 Emericella nidulans
66 771 Escherichia coli (strain K12 / DH10B)
67 765 Escherichia coli O127:H6 (strain E2348/69 / EPEC)
68 765 Escherichia coli (strain K12 / MC4100 / BW2952)
69 764 Escherichia coli O17:K52:H18 (strain UMN026 / ExPEC)
70 762 Escherichia coli (strain 55989 / EAEC)
71 761 Escherichia coli O8 (strain IAI1)
72 760 Shigella flexneri serotype 5b (strain 8401)
73 759 Staphylococcus epidermidis (strain ATCC 35984 / RP62A)
74 758 Staphylococcus epidermidis (strain ATCC 12228)
75 756 Escherichia coli (strain SE11)
76 756 Escherichia coli O45:K1 (strain S88 / ExPEC)
77 754 Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145)
78 753 Escherichia coli O7:K1 (strain IAI39 / ExPEC)
79 748 Escherichia coli O157:H7 (strain EC4115 / EHEC)
80 744 Photorhabdus luminescens subsp. laumondii (strain TT01)
81 740 Staphylococcus aureus (strain NCTC 8325)
82 735 Bacillus halodurans
83 735 Yersinia enterocolitica serotype O:8 / biotype 1B (strain 8081)
84 734 Bacillus anthracis
85 732 Vibrio vulnificus (strain CMCP6)
86 731 Escherichia coli O81 (strain ED1a)
87 721 Salmonella enteritidis PT4 (strain P125109)
88 717 Vibrio vulnificus (strain YJ016)
89 716 Salmonella paratyphi B (strain ATCC BAA-1250 / SPB7)
90 715 Yersinia pestis bv. Antiqua (strain Nepal516)
91 714 Salmonella paratyphi A (strain AKU_12601)
92 713 Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578)
93 713 Enterobacter sp. (strain 638)
94 713 Salmonella agona (strain SL483)
95 713 Escherichia coli O1:K1 / APEC
96 713 Salmonella newport (strain SL254)
97 713 Yersinia pseudotuberculosis serotype O:1b (strain IP 31758)
98 712 Salmonella schwarzengrund (strain CVM19633)
99 711 Yersinia pestis bv. Antiqua (strain Antiqua)
100 711 Zea mays (Maize)
101 710 Salmonella heidelberg (strain SL476)
102 702 Salmonella dublin (strain CT_02021853)
103 701 Candida albicans (strain SC5314 / ATCC MYA-2876) (Yeast)
104 698 Shigella boydii serotype 18 (strain CDC 3083-94 / BS512)
105 696 Klebsiella pneumoniae (strain 342)
106 695 Escherichia fergusonii (strain ATCC 35469 / DSM 13698 / CDC 0568-73)
107 693 Pseudomonas putida (strain KT2440)
108 689 Nostoc sp. (strain PCC 7120 / UTEX 2576)
109 688 Pan troglodytes (Chimpanzee)
110 687 Mycoplasma pneumoniae (strain ATCC 29342 / M129)
111 683 Salmonella gallinarum (strain 287/91 / NCTC 13346)
112 678 Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
113 675 Pseudomonas syringae pv. tomato (strain DC3000)
114 670 Serratia proteamaculans (strain 568)
115 668 Mycobacterium leprae (strain TN)
116 667 Yersinia pestis (strain Pestoides F)
117 666 Staphylococcus aureus (strain USA300)
118 658 Rhizobium sp. (strain NGR234)
119 654 Bradyrhizobium japonicum (strain USDA 110)
120 653 Debaryomyces hansenii
121 652 Bacillus cereus (strain ATCC 14579 / DSM 31)
122 650 Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100)
123 645 Escherichia coli
124 643 Staphylococcus aureus (strain bovine RF122 / ET3-1)
125 642 Salmonella arizonae (strain ATCC BAA-731 / CDC346-86 / RSK2980)
126 642 Yarrowia lipolytica (strain CLIB 122 / E 150) (Yeast) (Candida lipolytica)
127 638 Yersinia pseudotuberculosis serotype O:3 (strain YPIII)
128 634 Shewanella oneidensis (strain MR-1)
129 634 Yersinia pseudotuberculosis serotype IB (strain PB1/+)
130 632 Agrobacterium tumefaciens (strain C58 / ATCC 33970)
131 622 Cronobacter sakazakii (strain ATCC BAA-894) (Enterobacter sakazakii)
132 616 Treponema pallidum (strain Nichols)
133 615 Oryza sativa subsp. indica (Rice)
134 613 Methanothermobacter thermautotrophicus
135 612 Staphylococcus haemolyticus (strain JCSC1435)
136 606 Rhizobium loti (strain MAFF303099) (Mesorhizobium loti)
137 605 Xanthomonas campestris pv. campestris (strain ATCC 33913 / NCPPB 528 / LMG 568)
138 602 Ralstonia solanacearum (strain GMI1000) (Pseudomonas solanacearum)
139 602 Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e)
140 602 Photobacterium profundum (Photobacterium sp. (strain SS9))
141 602 Staphylococcus saprophyticus subsp. saprophyticus
142 601 Salmonella paratyphi C (strain RKS4594)
143 600 Yersinia pestis bv. Antiqua (strain Angola)
144 590 Bacillus cereus (strain ATCC 10987)
145 590 Listeria innocua serovar 6a (strain CLIP 11262)
146 589 Pectobacterium carotovorum subsp. carotovorum (strain PC1)
147 586 Rickettsia prowazekii (strain Madrid E)
148 580 Helicobacter pylori (strain ATCC 700392 / 26695) (Campylobacter pylori)
149 578 Neisseria meningitidis serogroup B (strain MC58)
150 576 Brucella suis biovar 1 (strain 1330)
151 572 Brucella melitensis biotype 1 (strain 16M / ATCC 23456 / NCTC 10094)
152 572 Buchnera aphidicola subsp. Acyrthosiphon pisum (strain APS)
153 568 Caenorhabditis briggsae
154 567 Bacillus thuringiensis subsp. konkukian (strain 97-27)
155 566 Pseudomonas syringae pv. syringae (strain B728a)
156 565 Helicobacter pylori (strain J99) (Campylobacter pylori J99)
157 564 Vibrio fischeri (strain ATCC 700601 / ES114)
158 564 Caulobacter crescentus (strain ATCC 19089 / CB15)
159 564 Pseudomonas aeruginosa (strain UCBPP-PA14)
160 562 Bacillus licheniformis (strain DSM 13 / ATCC 14580)
161 562 Buchnera aphidicola subsp. Schizaphis graminum (strain Sg)
162 561 Bacillus cereus (strain ZK / E33L)
163 556 Xanthomonas axonopodis pv. citri (strain 306)
164 556 Clostridium acetobutylicum
165 552 Oceanobacillus iheyensis (strain DSM 14371 / JCM 11309 / KCTC 3954 / HTE831)
166 552 Neisseria meningitidis serogroup A / serotype 4A (strain Z2491)
167 552 Pseudomonas fluorescens (strain Pf0-1)
168 546 Pseudomonas fluorescens (strain Pf-5 / ATCC BAA-477)
169 545 Pseudomonas syringae pv. phaseolicola (strain 1448A / Race 6)
170 533 Lactococcus lactis subsp. lactis (strain IL1403) (Streptococcus lactis)
171 531 Erwinia tasmaniensis (strain DSM 17950 / Et1/99)
172 529 Sodalis glossinidius (strain morsitans)
173 529 Listeria monocytogenes serotype 4b (strain F2365)
174 527 Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099)
175 522 Bordetella bronchiseptica (strain ATCC BAA-588 / NCTC 13252 / RB50)
176 522 Xylella fastidiosa (strain 9a5c)
177 515 Chromobacterium violaceum
178 515 Bordetella pertussis (strain Tohama I / ATCC BAA-589 / NCTC 13251)
179 512 Xylella fastidiosa (strain Temecula1 / ATCC 700964)
180 511 Pseudomonas aeruginosa (strain PA7)
181 511 Vibrio cholerae serotype O1 (strain ATCC 39541 / Ogawa 395 / O395)
182 510 Haemophilus ducreyi (strain 35000HP / ATCC 700724)
183 508 Bordetella parapertussis (strain 12822 / ATCC BAA-587 / NCTC 13253)
184 507 Buchnera aphidicola subsp. Baizongia pistaciae (strain Bp)
185 507 Geobacillus kaustophilus (strain HTA426)
186 506 Staphylococcus aureus (strain Newman)
187 502 Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4)
188 501 Deinococcus radiodurans
189 500 Pseudomonas entomophila (strain L48)
190 499 Corynebacterium glutamicum
191 499 Brucella abortus biovar 1 (strain 9-941)
192 497 Streptomyces avermitilis
193 497 Rickettsia conorii (strain ATCC VR-613 / Malish 7)
194 496 Bacillus clausii (strain KSM-K16)
195 495 Burkholderia pseudomallei (strain K96243)
196 495 Haemophilus influenzae (strain 86-028NP)
197 494 Proteus mirabilis (strain HI4320)
198 492 Bacillus amyloliquefaciens (strain FZB42)
199 491 Xanthomonas campestris pv. campestris (strain 8004)
200 490 Vibrio harveyi (strain ATCC BAA-1116 / BB120)
201 489 Methanosarcina acetivorans (strain ATCC 35395 / DSM 2834 / JCM 12185 / C2A)
202 487 Shewanella sp. (strain MR-7)
203 486 Mannheimia succiniciproducens (strain MBEL55E)
204 484 Pseudomonas aeruginosa (strain LESB58)
205 484 Staphylococcus aureus (strain Mu3 / ATCC 700698)
206 484 Shewanella sp. (strain MR-4)
207 483 Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195)
208 481 Thermosynechococcus elongatus (strain BP-1)
209 480 Acinetobacter sp. (strain ADP1)
210 479 Pyrococcus horikoshii
211 478 Synechococcus elongatus (strain PCC 7942) (Anacystis nidulans R2)
212 475 Pseudomonas putida (strain F1 / ATCC 700007)
213 474 Burkholderia sp. (strain 383) (Burkholderia cepacia
214 474 Brucella abortus (strain 2308)
215 473 Aspergillus oryzae (strain ATCC 42149 / RIB 40) (Yellow koji mold)
216 472 Streptococcus pneumoniae (strain ATCC BAA-255 / R6)
217 469 Methanosarcina mazei
218 467 Halobacterium salinarum (strain ATCC 700922 / JCM 11081 / NRC-1)
219 467 Clostridium perfringens (strain 13 / Type A)
220 466 Pyrococcus abyssi (strain GE5 / Orsay)
221 466 Xanthomonas campestris pv. vesicatoria (strain 85-10)
222 466 Shewanella frigidimarina (strain NCIMB 400)
223 466 Pseudomonas putida (strain GB-1)
224 466 Cupriavidus necator (strain ATCC 17699 / H16 / DSM 428 / Stanier 337)
225 464 Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / NCIB 9240)
226 463 Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1)
227 463 Shewanella sp. (strain ANA-3)
228 462 Burkholderia mallei (strain ATCC 23344)
229 462 Anabaena variabilis (strain ATCC 29413 / PCC 7937)
230 462 Rhodopseudomonas palustris (strain ATCC BAA-98 / CGA009)
231 461 Lactobacillus plantarum (strain ATCC BAA-793 / NCIMB 8826 / WCFS1)
232 459 Cupriavidus pinatubonensis (strain JMP134 / LMG 1197) (Alcaligenes eutrophus)
233 458 Enterococcus faecalis (strain ATCC 700802 / V583)
234 455 Staphylococcus aureus (strain JH1)
235 455 Rhodobacter sphaeroides (strain ATCC 17023 / 2.4.1 / NCIB 8253 / DSM 158)
236 454 Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath)
237 454 Xanthomonas oryzae pv. oryzae (strain MAFF 311018)
238 453 Ovis aries (Sheep)
239 453 Pseudomonas putida (strain W619)
240 453 Rickettsia felis (strain ATCC VR-1525 / URRWXCal2) (Rickettsia azadi)
241 452 Shewanella baltica (strain OS185)
242 451 Aeromonas salmonicida (strain A449)
243 449 Thermoanaerobacter tengcongensis
244 449 Staphylococcus aureus (strain JH9)
245 449 Hahella chejuensis (strain KCTC 2396)
246 448 Nicotiana tabacum (Common tobacco)
247 448 Mycobacterium paratuberculosis (strain ATCC BAA-968 / K-10)
248 448 Streptococcus mutans serotype c (strain ATCC 700610 / UA159)
249 447 Vibrio fischeri (strain MJ11)
250 446 Sulfolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2)
2.3 Taxonomic distribution of the sequences
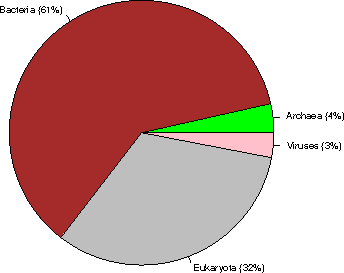 Kingdom sequences (% of the database)
Archaea 18953 ( 4%)
Bacteria 328550 ( 61%)
Eukaryota 174462 ( 32%)
Viruses 16294 ( 3%)
Within Eukaryota:
Kingdom sequences (% of the database)
Archaea 18953 ( 4%)
Bacteria 328550 ( 61%)
Eukaryota 174462 ( 32%)
Viruses 16294 ( 3%)
Within Eukaryota:
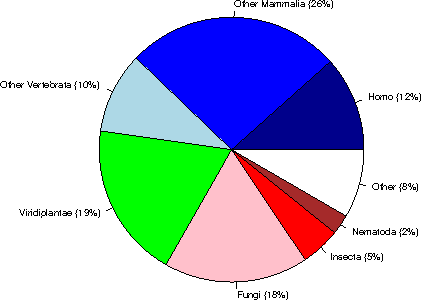 Category sequences (% of Eukaryota) (% of the complete database)
Human 20234 ( 12%) ( 4%)
Other Mammalia 45763 ( 26%) ( 9%)
Other Vertebrata 17270 ( 10%) ( 3%)
Viridiplantae 33169 ( 19%) ( 6%)
Fungi 30780 ( 18%) ( 6%)
Insecta 8425 ( 5%) ( 2%)
Nematoda 4238 ( 2%) ( 1%)
Other 14583 ( 8%) ( 3%)
3. SEQUENCE SIZE
Repartition of the sequences by size (excluding fragments)
From To Number From To Number
1- 50 8735 1001-1100 3733
51- 100 41281 1101-1200 2586
101- 150 57513 1201-1300 2009
151- 200 57638 1301-1400 1870
201- 250 56409 1401-1500 1502
251- 300 49836 1501-1600 725
301- 350 49996 1601-1700 562
351- 400 43196 1701-1800 459
401- 450 35392 1801-1900 423
451- 500 28489 1901-2000 349
501- 550 20245 2001-2100 212
551- 600 14523 2101-2200 283
601- 650 12217 2201-2300 292
651- 700 8818 2301-2400 172
701- 750 7262 2401-2500 136
751- 800 5159 >2500 1081
801- 850 4523
851- 900 5009
901- 950 3851
951-1000 2733
Category sequences (% of Eukaryota) (% of the complete database)
Human 20234 ( 12%) ( 4%)
Other Mammalia 45763 ( 26%) ( 9%)
Other Vertebrata 17270 ( 10%) ( 3%)
Viridiplantae 33169 ( 19%) ( 6%)
Fungi 30780 ( 18%) ( 6%)
Insecta 8425 ( 5%) ( 2%)
Nematoda 4238 ( 2%) ( 1%)
Other 14583 ( 8%) ( 3%)
3. SEQUENCE SIZE
Repartition of the sequences by size (excluding fragments)
From To Number From To Number
1- 50 8735 1001-1100 3733
51- 100 41281 1101-1200 2586
101- 150 57513 1201-1300 2009
151- 200 57638 1301-1400 1870
201- 250 56409 1401-1500 1502
251- 300 49836 1501-1600 725
301- 350 49996 1601-1700 562
351- 400 43196 1701-1800 459
401- 450 35392 1801-1900 423
451- 500 28489 1901-2000 349
501- 550 20245 2001-2100 212
551- 600 14523 2101-2200 283
601- 650 12217 2201-2300 292
651- 700 8818 2301-2400 172
701- 750 7262 2401-2500 136
751- 800 5159 >2500 1081
801- 850 4523
851- 900 5009
901- 950 3851
951-1000 2733
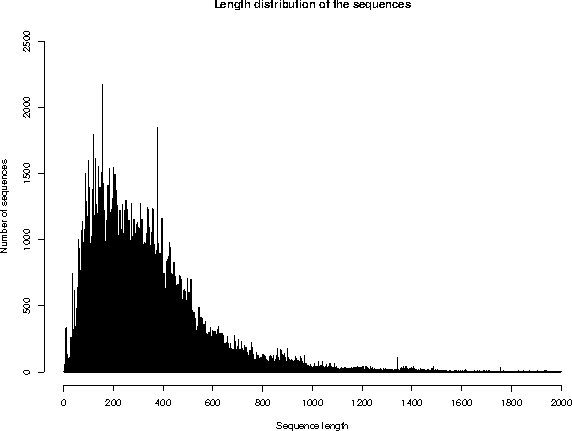 The average sequence length in UniProtKB/Swiss-Prot is 355 amino acids.
The shortest sequence is GWA_SEPOF (P83570): 2 amino acids.
The longest sequence is TITIN_MOUSE (A2ASS6): 35213 amino acids.
4. JOURNAL CITATIONS
Note: the following citation statistics reflect the number of distinct
journal citations.
Total number of journals cited in this release of UniProtKB/Swiss-Prot: 2280
4.1 Table of the frequency of journal citations
Journals cited 1x: 751
2x: 290
3x: 152
4x: 118
5x: 84
6x: 69
7x: 57
8x: 45
9x: 32
10x: 29
11- 20x: 182
21- 50x: 190
51-100x: 100
>100x: 181
4.2 List of the most cited journals in UniProtKB/Swiss-Prot
Nb Citations Journal name
-- --------- -------------------------------------------------------------
1 20577 Journal of Biological Chemistry
2 9326 Proceedings of the National Academy of Sciences of the U.S.A.
3 5604 Journal of Bacteriology
4 4973 Biochemical and Biophysical Research Communications
5 4585 Gene
6 4500 Nucleic Acids Research
7 4342 Biochemistry
8 4299 FEBS Letters
9 4155 The EMBO Journal
10 3868 Molecular and Cellular Biology
11 3636 Nature
12 3479 Journal of Molecular Biology
13 3226 European Journal of Biochemistry
14 3180 Biochimica et Biophysica Acta
15 2976 Cell
16 2503 Genomics
17 2463 Journal of Virology
18 2428 Biochemical Journal
19 2399 Science
20 1989 Molecular Microbiology
21 1822 Journal of Cell Biology
22 1769 Plant Physiology
23 1614 Plant Molecular Biology
24 1559 The American Journal of Human Genetics
25 1553 Genes and Development
26 1513 Virology
27 1476 Nature Genetics
28 1433 Human Molecular Genetics
29 1405 Oncogene
30 1331 Molecular and General Genetics
31 1304 Development
32 1290 Human Mutation
33 1255 Molecular Biology of the Cell
34 1225 The Plant Cell
35 1224 Journal of Biochemistry
36 1158 Journal of Immunology
37 1108 Molecular Cell
38 1095 The Plant Journal
39 1090 Genetics
40 1046 Structure
41 1013 Journal of General Virology
42 957 Blood
43 929 Infection and Immunity
44 918 Archives of Biochemistry and Biophysics
45 909 Journal of Cell Science
46 831 Microbiology
47 816 Developmental Biology
48 794 Cancer Research
49 783 Yeast
50 772 Current Biology
51 715 FEMS Microbiology Letters
52 649 Acta Crystallographica, Section D
53 636 Protein Science
54 629 Journal of Neuroscience
55 623 Applied and Environmental Microbiology
56 623 Toxicon
57 622 Human Genetics
58 618 Nature Structural Biology
59 616 Mechanisms of Development
60 579 Neuron
61 565 Journal of Clinical Investigation
62 537 Current Genetics
63 532 American Journal of Physiology
64 522 The Journal of Experimental Medicine
65 483 Proteins
66 481 Molecular Endocrinology
67 479 Mammalian Genome
68 457 Journal of Neurochemistry
69 457 PLoS ONE
70 456 Immunogenetics
71 441 The Journal of Clinical Endocrinology and Metabolism
72 441 Plant and Cell Physiology
73 433 Bioscience, Biotechnology, and Biochemistry
74 430 Endocrinology
75 428 Molecular and Biochemical Parasitology
76 423 Nature Cell Biology
77 400 Journal of Medical Genetics
78 395 Journal of Molecular Evolution
79 377 DNA and Cell Biology
80 375 Molecular Biology and Evolution
81 367 Experimental Cell Research
82 364 DNA Sequence
83 342 Peptides
84 326 Brain Research. Molecular Brain Research
85 324 Tissue Antigens
86 318 Comparative Biochemistry and Physiology
87 311 Developmental Cell
88 310 RNA
89 310 Antimicrobial Agents and Chemotherapy
90 303 Journal of Investigative Dermatology
91 302 Molecular Pharmacology
92 294 Biological Chemistry Hoppe-Seyler
93 293 Nature Structural and Molecular Biology
94 290 Planta
95 288 The FEBS Journal
96 280 Biology of Reproduction
97 279 Cytogenetics and Cell Genetics
98 277 Neurology
99 266 Developmental Dynamics
100 263 Genome Research
101 262 Virus Research
102 254 Journal of General Microbiology
103 248 EMBO Reports
104 245 Immunity
105 243 Molecular Plant-Microbe Interactions
106 243 Biochimie
107 235 Eukaryotic Cell
108 233 The New England Journal of Medicine
109 232 The FASEB Journal
110 232 Genes to Cells
111 227 European Journal of Immunology
112 222 Annals of Neurology
113 219 European Journal of Human Genetics
114 218 Hoppe-Seyler's Zeitschrift fur Physiologische Chemie
115 212 DNA Research
116 209 Journal of Human Genetics
117 197 Investigative Ophthalmology and Visual Science
118 196 Archives of Microbiology
119 195 Journal of the American Chemical Society
120 188 Nature Immunology
121 188 Archives of Virology
122 184 Journal of Cellular Biochemistry
123 183 Molecular and Cellular Endocrinology
124 183 American Journal of Medical Genetics. Part A
125 182 BMC Genomics
126 180 Molecular Immunology
127 177 Glycobiology
128 176 Clinical Genetics
129 172 Insect Biochemistry and Molecular Biology
130 171 Diabetes
131 168 American Journal of Medical Genetics
132 167 Journal of Medicinal Chemistry
133 167 Molecular Phylogenetics and Evolution
134 165 Acta Crystallographica, Section F
135 165 Journal of Experimental Botany
136 163 Circulation Research
137 159 International Journal of Cancer
138 159 DNA
139 158 Molecular Reproduction and Development
140 155 Hemoglobin
141 154 Bioorganicheskaia Khimiia
142 154 Molecular Genetics and Metabolism
143 150 PLoS Genetics
144 149 Phytochemistry
145 149 Molecular and Cellular Neuroscience
146 149 Biological Chemistry
147 148 Molecular Genetics and Genomics
148 145 Journal of Lipid Research
149 145 Protein Expression and Purification
150 144 Traffic
5. STATISTICS FOR SOME LINE TYPES
The following table summarizes the total number of some UniProtKB/Swiss-Prot lines,
as well as the number of entries with at least one such line, and the
frequency of the lines.
Total Number of Average
Line type / subtype number entries per entry
------------------------------------ -------- --------- ---------
References (RL) 1028777 1.91
Journal 822062 413904 1.53 1
Submitted to EMBL/GenBank/DDBJ 197677 177199 0.37 2
Submitted to other databases 6851 6388 0.01 3
Book citation 717 703 <0.01 4
Plant Gene Register 578 566 <0.01 5
Thesis 413 410 <0.01 6
Unpublished observations 283 279 <0.01 7
Patent 190 187 <0.01 8
Worm Breeder's Gazette 6 6 <0.01 9
Total number of distinct authors cited in UniProtKB/Swiss-Prot: 328522
Total Number of Average
Line type / subtype number entries per entry Rank
------------------------------------ -------- --------- --------- ----
Comments (CC) 2400283 4.46
ALLERGEN 520 520 <0.01 26
ALTERNATIVE PRODUCTS 21332 21332 0.04 13
BIOPHYSICOCHEMICAL PROPERTIES 4624 4624 0.01 23
BIOTECHNOLOGY 332 330 <0.01 28
CATALYTIC ACTIVITY 242353 218947 0.45 4
CAUTION 8087 7921 0.02 19
COFACTOR 106419 97309 0.20 7
DEVELOPMENTAL STAGE 9691 9691 0.02 16
DISEASE 5134 3440 0.01 21
DISRUPTION PHENOTYPE 5045 5045 0.01 22
DOMAIN 38259 33859 0.07 10
ENZYME REGULATION 10841 10841 0.02 15
FUNCTION 416405 399296 0.77 2
INDUCTION 14616 14616 0.03 14
INTERACTION 9601 9601 0.02 17
MASS SPECTROMETRY 5343 4073 0.01 20
MISCELLANEOUS 32311 29870 0.06 12
PATHWAY 131430 119277 0.24 6
PHARMACEUTICAL 85 85 <0.01 29
POLYMORPHISM 876 823 <0.01 24
PTM 44519 35135 0.08 8
RNA EDITING 623 623 <0.01 25
SEQUENCE CAUTION 40704 40704 0.08 9
SIMILARITY 638064 513536 1.19 1
SUBCELLULAR LOCATION 324857 319065 0.60 3
SUBUNIT 240928 240928 0.45 5
TISSUE SPECIFICITY 37920 37920 0.07 11
TOXIC DOSE 510 495 <0.01 27
WEB RESOURCE 8854 7084 0.02 18
Total number of comment topics: 29
Total Number of Average
Line type / subtype number entries per entry Rank
------------------------------------ -------- --------- --------- ----
Features (FT) 3667028 6.81
ACT_SITE 139190 85292 0.26 9
BINDING 272605 73809 0.51 4
CA_BIND 3834 1587 0.01 35
CARBOHYD 106764 27346 0.20 15
CHAIN 545155 532346 1.01 1
COILED 20000 13732 0.04 26
COMPBIAS 53963 28565 0.10 18
CONFLICT 126255 44288 0.23 11
CROSSLNK 6614 3859 0.01 34
DISULFID 107396 29175 0.20 14
DNA_BIND 10286 9398 0.02 31
DOMAIN 159725 94793 0.30 8
HELIX 173302 16986 0.32 7
INIT_MET 15301 15301 0.03 27
INTRAMEM 2139 919 <0.01 37
LIPID 11831 7499 0.02 30
METAL 310967 76643 0.58 3
MOD_RES 194070 63478 0.36 5
MOTIF 35440 22924 0.07 24
MUTAGEN 41260 9610 0.08 22
NON_CONS 2028 743 <0.01 38
NON_STD 354 279 <0.01 39
NON_TER 12186 9309 0.02 29
NP_BIND 116169 72454 0.22 13
PEPTIDE 9908 6692 0.02 32
PROPEP 12583 10817 0.02 28
REGION 126210 65168 0.23 12
REPEAT 94031 13911 0.17 16
SIGNAL 37983 37973 0.07 23
SITE 42620 24644 0.08 20
STRAND 181967 16059 0.34 6
TOPO_DOM 129335 26817 0.24 10
TRANSIT 8193 8090 0.02 33
TRANSMEM 354653 73165 0.66 2
TURN 42316 13753 0.08 21
UNSURE 3087 573 0.01 36
VAR_SEQ 43052 18503 0.08 19
VARIANT 84974 16695 0.16 17
ZN_FING 29282 12882 0.05 25
Total number of feature keys: 39
Total Number of Average
Line type / subtype number entries per entry Rank Category
------------------------------------ -------- --------- --------- ---- -------------------------------------------
Cross-references (DR) 15576158 28.94
2DBase-Ecoli 85 85 <0.01 127 2D gel databases
Aarhus/Ghent-2DPAGE 126 96 <0.01 124 2D gel databases
AGD 932 926 <0.01 102 Organism-specific databases
Allergome 1446 897 <0.01 98 Protein family/group databases
ANU-2DPAGE 27 27 <0.01 134 2D gel databases
ArachnoServer 763 755 <0.01 108 Organism-specific databases
ArrayExpress 59926 59926 0.11 43 Gene expression databases
Bgee 39199 39199 0.07 46 Gene expression databases
BindingDB 295 295 <0.01 120 Other
BioCyc 248606 240153 0.46 22 Enzyme and pathway databases
BRENDA 4294 4285 0.01 89 Enzyme and pathway databases
CAZy 7581 6822 0.01 74 Protein family/group databases
CGD 682 660 <0.01 109 Organism-specific databases
CleanEx 30094 29453 0.06 51 Gene expression databases
COMPLUYEAST-2DPAGE 99 98 <0.01 126 2D gel databases
ConoServer 915 833 <0.01 104 Organism-specific databases
Cornea-2DPAGE 67 67 <0.01 128 2D gel databases
CTD 69075 68422 0.13 41 Organism-specific databases
CYGD 5596 5593 0.01 78 Organism-specific databases
dictyBase 4203 4086 0.01 91 Organism-specific databases
DIP 13810 13714 0.03 68 Protein-protein interaction databases
DisProt 398 395 <0.01 117 3D structure databases
DMDM 16765 16762 0.03 64 Polymorphism databases
DNASU 18658 18587 0.03 59 Protocols and materials databases
DOSAC-COBS-2DPAGE 149 147 <0.01 123 2D gel databases
DrugBank 5319 1628 0.01 80 Other
EchoBASE 4167 4163 0.01 92 Organism-specific databases
ECO2DBASE 352 300 <0.01 119 2D gel databases
EcoGene 4292 4290 0.01 90 Organism-specific databases
eggNOG 430090 430090 0.80 9 Phylogenomic databases
EMBL 929954 527394 1.73 3 Sequence databases
Ensembl 79881 48017 0.15 35 Genome annotation databases
EnsemblBacteria 97993 85019 0.18 31 Genome annotation databases
EnsemblFungi 17419 17127 0.03 63 Genome annotation databases
EnsemblMetazoa 11598 8843 0.02 70 Genome annotation databases
EnsemblPlants 15117 13392 0.03 67 Genome annotation databases
EnsemblProtists 4462 4337 0.01 88 Genome annotation databases
euHCVdb 55 44 <0.01 129 Organism-specific databases
EuPathDB 808 807 <0.01 105 Organism-specific databases
EvolutionaryTrace 16512 16512 0.03 65 Other
FlyBase 5864 5490 0.01 77 Organism-specific databases
Gene3D 330441 253538 0.61 17 Family and domain databases
GeneCards 20037 19729 0.04 56 Organism-specific databases
GeneFarm 3078 3067 0.01 94 Organism-specific databases
GeneID 498069 469564 0.93 6 Genome annotation databases
GeneTree 34526 34511 0.06 47 Phylogenomic databases
Genevestigator 66904 66904 0.12 42 Gene expression databases
GenoList 7069 7057 0.01 75 Organism-specific databases
GenomeReviews 377158 357543 0.70 13 Genome annotation databases
GenomeRNAi 21203 21203 0.04 54 Other
GermOnline 41901 41327 0.08 45 Gene expression databases
GlycoSuiteDB 272 272 <0.01 121 PTM databases
GO 2350326 507710 4.37 1 Ontologies
Gramene 4853 4853 0.01 83 Organism-specific databases
H-InvDB 5596 4774 0.01 79 Organism-specific databases
HAMAP 312264 312058 0.58 18 Family and domain databases
HGNC 19815 19656 0.04 57 Organism-specific databases
HOGENOM 384625 384625 0.71 12 Phylogenomic databases
HOVERGEN 75477 75477 0.14 38 Phylogenomic databases
HPA 17640 13724 0.03 61 Organism-specific databases
HSSP 30466 30466 0.06 50 3D structure databases
InParanoid 69470 69470 0.13 40 Phylogenomic databases
IntAct 33930 33930 0.06 48 Protein-protein interaction databases
InterPro 1769886 514871 3.29 2 Family and domain databases
IPI 95378 67175 0.18 32 Sequence databases
KEGG 473526 447533 0.88 8 Genome annotation databases
KO 371542 371157 0.69 14 Phylogenomic databases
LegioList 765 763 <0.01 107 Organism-specific databases
Leproma 671 668 <0.01 110 Organism-specific databases
MaizeGDB 496 491 <0.01 115 Organism-specific databases
MEROPS 10484 10484 0.02 72 Protein family/group databases
MGI 16477 16432 0.03 66 Organism-specific databases
MIM 17821 13538 0.03 60 Organism-specific databases
MINT 17619 17619 0.03 62 Protein-protein interaction databases
NextBio 69701 69701 0.13 39 Other
neXtProt 20084 20084 0.04 55 Organism-specific databases
OGP 377 377 <0.01 118 2D gel databases
OMA 390637 390637 0.73 11 Phylogenomic databases
Orphanet 4478 2566 0.01 87 Organism-specific databases
OrthoDB 78128 78128 0.15 37 Phylogenomic databases
PANTHER 191286 178861 0.36 24 Family and domain databases
Pathway_Interaction_DB 4568 1666 0.01 86 Enzyme and pathway databases
PATRIC 308720 308695 0.57 20 Genome annotation databases
PDB 89369 18697 0.17 34 3D structure databases
PDBsum 89369 18697 0.17 33 3D structure databases
PeptideAtlas 5163 5163 0.01 81 Proteomic databases
PeroxiBase 768 752 <0.01 106 Protein family/group databases
Pfam 711684 495206 1.32 4 Family and domain databases
PharmGKB 18758 18341 0.03 58 Organism-specific databases
PHCI-2DPAGE 250 250 <0.01 122 2D gel databases
PhosphoSite 33567 33567 0.06 49 PTM databases
PhosSite 627 615 <0.01 111 PTM databases
PhylomeDB 26846 26846 0.05 53 Phylogenomic databases
PIR 118224 108103 0.22 28 Sequence databases
PIRSF 98487 98472 0.18 29 Family and domain databases
PMAP-CutDB 1457 1457 <0.01 97 Other
PMMA-2DPAGE 52 52 <0.01 130 2D gel databases
PomBase 5016 4958 0.01 82 Organism-specific databases
PptaseDB 38 38 <0.01 131 Protein family/group databases
PRIDE 79833 79833 0.15 36 Proteomic databases
PRINTS 137353 120147 0.26 26 Family and domain databases
ProDom 29293 29114 0.05 52 Family and domain databases
ProMEX 513 513 <0.01 113 Proteomic databases
PROSITE 480044 303359 0.89 7 Family and domain databases
ProtClustDB 343244 343244 0.64 15 Phylogenomic databases
ProteinModelPortal 429127 429127 0.80 10 3D structure databases
PseudoCAP 1250 1241 <0.01 100 Organism-specific databases
Rat-heart-2DPAGE 28 28 <0.01 133 2D gel databases
Reactome 12221 7551 0.02 69 Enzyme and pathway databases
REBASE 402 402 <0.01 116 Protein family/group databases
RefSeq 521452 469836 0.97 5 Sequence databases
REPRODUCTION-2DPAGE 1256 1035 <0.01 99 2D gel databases
RGD 7724 7720 0.01 73 Organism-specific databases
SGD 6640 6635 0.01 76 Organism-specific databases
Siena-2DPAGE 103 103 <0.01 125 2D gel databases
SMART 167200 125162 0.31 25 Family and domain databases
SMR 218856 218856 0.41 23 3D structure databases
STRING 309313 309310 0.57 19 Protein-protein interaction databases
SUPFAM 330871 261981 0.61 16 Family and domain databases
SWISS-2DPAGE 1182 1181 <0.01 101 2D gel databases
TAIR 11588 11535 0.02 71 Organism-specific databases
TCDB 3647 3632 0.01 93 Protein family/group databases
TIGR 35 33 <0.01 132 Genome annotation databases
TIGRFAMs 289887 269320 0.54 21 Family and domain databases
TubercuList 1982 1946 <0.01 96 Organism-specific databases
UCD-2DPAGE 509 500 <0.01 114 2D gel databases
UCSC 51926 39791 0.10 44 Genome annotation databases
UniGene 98447 89672 0.18 30 Sequence databases
UniPathway 131304 119154 0.24 27 Enzyme and pathway databases
VectorBase 606 588 <0.01 112 Genome annotation databases
World-2DPAGE 919 908 <0.01 103 2D gel databases
WormBase 4798 3880 0.01 84 Organism-specific databases
Xenbase 4717 4712 0.01 85 Organism-specific databases
ZFIN 2775 2775 0.01 95 Organism-specific databases
Total number of cross-referenced databases: 134
6. AMINO ACID COMPOSITION
6.1 Composition in percent for the complete database
Ala (A) 8.25 Gln (Q) 3.93 Leu (L) 9.66 Ser (S) 6.56
Arg (R) 5.53 Glu (E) 6.75 Lys (K) 5.84 Thr (T) 5.34
Asn (N) 4.06 Gly (G) 7.08 Met (M) 2.42 Trp (W) 1.08
Asp (D) 5.45 His (H) 2.27 Phe (F) 3.86 Tyr (Y) 2.92
Cys (C) 1.37 Ile (I) 5.96 Pro (P) 4.70 Val (V) 6.87
Asx (B) 0.000 Glx (Z) 0.000 Xaa (X) 0.00
The average sequence length in UniProtKB/Swiss-Prot is 355 amino acids.
The shortest sequence is GWA_SEPOF (P83570): 2 amino acids.
The longest sequence is TITIN_MOUSE (A2ASS6): 35213 amino acids.
4. JOURNAL CITATIONS
Note: the following citation statistics reflect the number of distinct
journal citations.
Total number of journals cited in this release of UniProtKB/Swiss-Prot: 2280
4.1 Table of the frequency of journal citations
Journals cited 1x: 751
2x: 290
3x: 152
4x: 118
5x: 84
6x: 69
7x: 57
8x: 45
9x: 32
10x: 29
11- 20x: 182
21- 50x: 190
51-100x: 100
>100x: 181
4.2 List of the most cited journals in UniProtKB/Swiss-Prot
Nb Citations Journal name
-- --------- -------------------------------------------------------------
1 20577 Journal of Biological Chemistry
2 9326 Proceedings of the National Academy of Sciences of the U.S.A.
3 5604 Journal of Bacteriology
4 4973 Biochemical and Biophysical Research Communications
5 4585 Gene
6 4500 Nucleic Acids Research
7 4342 Biochemistry
8 4299 FEBS Letters
9 4155 The EMBO Journal
10 3868 Molecular and Cellular Biology
11 3636 Nature
12 3479 Journal of Molecular Biology
13 3226 European Journal of Biochemistry
14 3180 Biochimica et Biophysica Acta
15 2976 Cell
16 2503 Genomics
17 2463 Journal of Virology
18 2428 Biochemical Journal
19 2399 Science
20 1989 Molecular Microbiology
21 1822 Journal of Cell Biology
22 1769 Plant Physiology
23 1614 Plant Molecular Biology
24 1559 The American Journal of Human Genetics
25 1553 Genes and Development
26 1513 Virology
27 1476 Nature Genetics
28 1433 Human Molecular Genetics
29 1405 Oncogene
30 1331 Molecular and General Genetics
31 1304 Development
32 1290 Human Mutation
33 1255 Molecular Biology of the Cell
34 1225 The Plant Cell
35 1224 Journal of Biochemistry
36 1158 Journal of Immunology
37 1108 Molecular Cell
38 1095 The Plant Journal
39 1090 Genetics
40 1046 Structure
41 1013 Journal of General Virology
42 957 Blood
43 929 Infection and Immunity
44 918 Archives of Biochemistry and Biophysics
45 909 Journal of Cell Science
46 831 Microbiology
47 816 Developmental Biology
48 794 Cancer Research
49 783 Yeast
50 772 Current Biology
51 715 FEMS Microbiology Letters
52 649 Acta Crystallographica, Section D
53 636 Protein Science
54 629 Journal of Neuroscience
55 623 Applied and Environmental Microbiology
56 623 Toxicon
57 622 Human Genetics
58 618 Nature Structural Biology
59 616 Mechanisms of Development
60 579 Neuron
61 565 Journal of Clinical Investigation
62 537 Current Genetics
63 532 American Journal of Physiology
64 522 The Journal of Experimental Medicine
65 483 Proteins
66 481 Molecular Endocrinology
67 479 Mammalian Genome
68 457 Journal of Neurochemistry
69 457 PLoS ONE
70 456 Immunogenetics
71 441 The Journal of Clinical Endocrinology and Metabolism
72 441 Plant and Cell Physiology
73 433 Bioscience, Biotechnology, and Biochemistry
74 430 Endocrinology
75 428 Molecular and Biochemical Parasitology
76 423 Nature Cell Biology
77 400 Journal of Medical Genetics
78 395 Journal of Molecular Evolution
79 377 DNA and Cell Biology
80 375 Molecular Biology and Evolution
81 367 Experimental Cell Research
82 364 DNA Sequence
83 342 Peptides
84 326 Brain Research. Molecular Brain Research
85 324 Tissue Antigens
86 318 Comparative Biochemistry and Physiology
87 311 Developmental Cell
88 310 RNA
89 310 Antimicrobial Agents and Chemotherapy
90 303 Journal of Investigative Dermatology
91 302 Molecular Pharmacology
92 294 Biological Chemistry Hoppe-Seyler
93 293 Nature Structural and Molecular Biology
94 290 Planta
95 288 The FEBS Journal
96 280 Biology of Reproduction
97 279 Cytogenetics and Cell Genetics
98 277 Neurology
99 266 Developmental Dynamics
100 263 Genome Research
101 262 Virus Research
102 254 Journal of General Microbiology
103 248 EMBO Reports
104 245 Immunity
105 243 Molecular Plant-Microbe Interactions
106 243 Biochimie
107 235 Eukaryotic Cell
108 233 The New England Journal of Medicine
109 232 The FASEB Journal
110 232 Genes to Cells
111 227 European Journal of Immunology
112 222 Annals of Neurology
113 219 European Journal of Human Genetics
114 218 Hoppe-Seyler's Zeitschrift fur Physiologische Chemie
115 212 DNA Research
116 209 Journal of Human Genetics
117 197 Investigative Ophthalmology and Visual Science
118 196 Archives of Microbiology
119 195 Journal of the American Chemical Society
120 188 Nature Immunology
121 188 Archives of Virology
122 184 Journal of Cellular Biochemistry
123 183 Molecular and Cellular Endocrinology
124 183 American Journal of Medical Genetics. Part A
125 182 BMC Genomics
126 180 Molecular Immunology
127 177 Glycobiology
128 176 Clinical Genetics
129 172 Insect Biochemistry and Molecular Biology
130 171 Diabetes
131 168 American Journal of Medical Genetics
132 167 Journal of Medicinal Chemistry
133 167 Molecular Phylogenetics and Evolution
134 165 Acta Crystallographica, Section F
135 165 Journal of Experimental Botany
136 163 Circulation Research
137 159 International Journal of Cancer
138 159 DNA
139 158 Molecular Reproduction and Development
140 155 Hemoglobin
141 154 Bioorganicheskaia Khimiia
142 154 Molecular Genetics and Metabolism
143 150 PLoS Genetics
144 149 Phytochemistry
145 149 Molecular and Cellular Neuroscience
146 149 Biological Chemistry
147 148 Molecular Genetics and Genomics
148 145 Journal of Lipid Research
149 145 Protein Expression and Purification
150 144 Traffic
5. STATISTICS FOR SOME LINE TYPES
The following table summarizes the total number of some UniProtKB/Swiss-Prot lines,
as well as the number of entries with at least one such line, and the
frequency of the lines.
Total Number of Average
Line type / subtype number entries per entry
------------------------------------ -------- --------- ---------
References (RL) 1028777 1.91
Journal 822062 413904 1.53 1
Submitted to EMBL/GenBank/DDBJ 197677 177199 0.37 2
Submitted to other databases 6851 6388 0.01 3
Book citation 717 703 <0.01 4
Plant Gene Register 578 566 <0.01 5
Thesis 413 410 <0.01 6
Unpublished observations 283 279 <0.01 7
Patent 190 187 <0.01 8
Worm Breeder's Gazette 6 6 <0.01 9
Total number of distinct authors cited in UniProtKB/Swiss-Prot: 328522
Total Number of Average
Line type / subtype number entries per entry Rank
------------------------------------ -------- --------- --------- ----
Comments (CC) 2400283 4.46
ALLERGEN 520 520 <0.01 26
ALTERNATIVE PRODUCTS 21332 21332 0.04 13
BIOPHYSICOCHEMICAL PROPERTIES 4624 4624 0.01 23
BIOTECHNOLOGY 332 330 <0.01 28
CATALYTIC ACTIVITY 242353 218947 0.45 4
CAUTION 8087 7921 0.02 19
COFACTOR 106419 97309 0.20 7
DEVELOPMENTAL STAGE 9691 9691 0.02 16
DISEASE 5134 3440 0.01 21
DISRUPTION PHENOTYPE 5045 5045 0.01 22
DOMAIN 38259 33859 0.07 10
ENZYME REGULATION 10841 10841 0.02 15
FUNCTION 416405 399296 0.77 2
INDUCTION 14616 14616 0.03 14
INTERACTION 9601 9601 0.02 17
MASS SPECTROMETRY 5343 4073 0.01 20
MISCELLANEOUS 32311 29870 0.06 12
PATHWAY 131430 119277 0.24 6
PHARMACEUTICAL 85 85 <0.01 29
POLYMORPHISM 876 823 <0.01 24
PTM 44519 35135 0.08 8
RNA EDITING 623 623 <0.01 25
SEQUENCE CAUTION 40704 40704 0.08 9
SIMILARITY 638064 513536 1.19 1
SUBCELLULAR LOCATION 324857 319065 0.60 3
SUBUNIT 240928 240928 0.45 5
TISSUE SPECIFICITY 37920 37920 0.07 11
TOXIC DOSE 510 495 <0.01 27
WEB RESOURCE 8854 7084 0.02 18
Total number of comment topics: 29
Total Number of Average
Line type / subtype number entries per entry Rank
------------------------------------ -------- --------- --------- ----
Features (FT) 3667028 6.81
ACT_SITE 139190 85292 0.26 9
BINDING 272605 73809 0.51 4
CA_BIND 3834 1587 0.01 35
CARBOHYD 106764 27346 0.20 15
CHAIN 545155 532346 1.01 1
COILED 20000 13732 0.04 26
COMPBIAS 53963 28565 0.10 18
CONFLICT 126255 44288 0.23 11
CROSSLNK 6614 3859 0.01 34
DISULFID 107396 29175 0.20 14
DNA_BIND 10286 9398 0.02 31
DOMAIN 159725 94793 0.30 8
HELIX 173302 16986 0.32 7
INIT_MET 15301 15301 0.03 27
INTRAMEM 2139 919 <0.01 37
LIPID 11831 7499 0.02 30
METAL 310967 76643 0.58 3
MOD_RES 194070 63478 0.36 5
MOTIF 35440 22924 0.07 24
MUTAGEN 41260 9610 0.08 22
NON_CONS 2028 743 <0.01 38
NON_STD 354 279 <0.01 39
NON_TER 12186 9309 0.02 29
NP_BIND 116169 72454 0.22 13
PEPTIDE 9908 6692 0.02 32
PROPEP 12583 10817 0.02 28
REGION 126210 65168 0.23 12
REPEAT 94031 13911 0.17 16
SIGNAL 37983 37973 0.07 23
SITE 42620 24644 0.08 20
STRAND 181967 16059 0.34 6
TOPO_DOM 129335 26817 0.24 10
TRANSIT 8193 8090 0.02 33
TRANSMEM 354653 73165 0.66 2
TURN 42316 13753 0.08 21
UNSURE 3087 573 0.01 36
VAR_SEQ 43052 18503 0.08 19
VARIANT 84974 16695 0.16 17
ZN_FING 29282 12882 0.05 25
Total number of feature keys: 39
Total Number of Average
Line type / subtype number entries per entry Rank Category
------------------------------------ -------- --------- --------- ---- -------------------------------------------
Cross-references (DR) 15576158 28.94
2DBase-Ecoli 85 85 <0.01 127 2D gel databases
Aarhus/Ghent-2DPAGE 126 96 <0.01 124 2D gel databases
AGD 932 926 <0.01 102 Organism-specific databases
Allergome 1446 897 <0.01 98 Protein family/group databases
ANU-2DPAGE 27 27 <0.01 134 2D gel databases
ArachnoServer 763 755 <0.01 108 Organism-specific databases
ArrayExpress 59926 59926 0.11 43 Gene expression databases
Bgee 39199 39199 0.07 46 Gene expression databases
BindingDB 295 295 <0.01 120 Other
BioCyc 248606 240153 0.46 22 Enzyme and pathway databases
BRENDA 4294 4285 0.01 89 Enzyme and pathway databases
CAZy 7581 6822 0.01 74 Protein family/group databases
CGD 682 660 <0.01 109 Organism-specific databases
CleanEx 30094 29453 0.06 51 Gene expression databases
COMPLUYEAST-2DPAGE 99 98 <0.01 126 2D gel databases
ConoServer 915 833 <0.01 104 Organism-specific databases
Cornea-2DPAGE 67 67 <0.01 128 2D gel databases
CTD 69075 68422 0.13 41 Organism-specific databases
CYGD 5596 5593 0.01 78 Organism-specific databases
dictyBase 4203 4086 0.01 91 Organism-specific databases
DIP 13810 13714 0.03 68 Protein-protein interaction databases
DisProt 398 395 <0.01 117 3D structure databases
DMDM 16765 16762 0.03 64 Polymorphism databases
DNASU 18658 18587 0.03 59 Protocols and materials databases
DOSAC-COBS-2DPAGE 149 147 <0.01 123 2D gel databases
DrugBank 5319 1628 0.01 80 Other
EchoBASE 4167 4163 0.01 92 Organism-specific databases
ECO2DBASE 352 300 <0.01 119 2D gel databases
EcoGene 4292 4290 0.01 90 Organism-specific databases
eggNOG 430090 430090 0.80 9 Phylogenomic databases
EMBL 929954 527394 1.73 3 Sequence databases
Ensembl 79881 48017 0.15 35 Genome annotation databases
EnsemblBacteria 97993 85019 0.18 31 Genome annotation databases
EnsemblFungi 17419 17127 0.03 63 Genome annotation databases
EnsemblMetazoa 11598 8843 0.02 70 Genome annotation databases
EnsemblPlants 15117 13392 0.03 67 Genome annotation databases
EnsemblProtists 4462 4337 0.01 88 Genome annotation databases
euHCVdb 55 44 <0.01 129 Organism-specific databases
EuPathDB 808 807 <0.01 105 Organism-specific databases
EvolutionaryTrace 16512 16512 0.03 65 Other
FlyBase 5864 5490 0.01 77 Organism-specific databases
Gene3D 330441 253538 0.61 17 Family and domain databases
GeneCards 20037 19729 0.04 56 Organism-specific databases
GeneFarm 3078 3067 0.01 94 Organism-specific databases
GeneID 498069 469564 0.93 6 Genome annotation databases
GeneTree 34526 34511 0.06 47 Phylogenomic databases
Genevestigator 66904 66904 0.12 42 Gene expression databases
GenoList 7069 7057 0.01 75 Organism-specific databases
GenomeReviews 377158 357543 0.70 13 Genome annotation databases
GenomeRNAi 21203 21203 0.04 54 Other
GermOnline 41901 41327 0.08 45 Gene expression databases
GlycoSuiteDB 272 272 <0.01 121 PTM databases
GO 2350326 507710 4.37 1 Ontologies
Gramene 4853 4853 0.01 83 Organism-specific databases
H-InvDB 5596 4774 0.01 79 Organism-specific databases
HAMAP 312264 312058 0.58 18 Family and domain databases
HGNC 19815 19656 0.04 57 Organism-specific databases
HOGENOM 384625 384625 0.71 12 Phylogenomic databases
HOVERGEN 75477 75477 0.14 38 Phylogenomic databases
HPA 17640 13724 0.03 61 Organism-specific databases
HSSP 30466 30466 0.06 50 3D structure databases
InParanoid 69470 69470 0.13 40 Phylogenomic databases
IntAct 33930 33930 0.06 48 Protein-protein interaction databases
InterPro 1769886 514871 3.29 2 Family and domain databases
IPI 95378 67175 0.18 32 Sequence databases
KEGG 473526 447533 0.88 8 Genome annotation databases
KO 371542 371157 0.69 14 Phylogenomic databases
LegioList 765 763 <0.01 107 Organism-specific databases
Leproma 671 668 <0.01 110 Organism-specific databases
MaizeGDB 496 491 <0.01 115 Organism-specific databases
MEROPS 10484 10484 0.02 72 Protein family/group databases
MGI 16477 16432 0.03 66 Organism-specific databases
MIM 17821 13538 0.03 60 Organism-specific databases
MINT 17619 17619 0.03 62 Protein-protein interaction databases
NextBio 69701 69701 0.13 39 Other
neXtProt 20084 20084 0.04 55 Organism-specific databases
OGP 377 377 <0.01 118 2D gel databases
OMA 390637 390637 0.73 11 Phylogenomic databases
Orphanet 4478 2566 0.01 87 Organism-specific databases
OrthoDB 78128 78128 0.15 37 Phylogenomic databases
PANTHER 191286 178861 0.36 24 Family and domain databases
Pathway_Interaction_DB 4568 1666 0.01 86 Enzyme and pathway databases
PATRIC 308720 308695 0.57 20 Genome annotation databases
PDB 89369 18697 0.17 34 3D structure databases
PDBsum 89369 18697 0.17 33 3D structure databases
PeptideAtlas 5163 5163 0.01 81 Proteomic databases
PeroxiBase 768 752 <0.01 106 Protein family/group databases
Pfam 711684 495206 1.32 4 Family and domain databases
PharmGKB 18758 18341 0.03 58 Organism-specific databases
PHCI-2DPAGE 250 250 <0.01 122 2D gel databases
PhosphoSite 33567 33567 0.06 49 PTM databases
PhosSite 627 615 <0.01 111 PTM databases
PhylomeDB 26846 26846 0.05 53 Phylogenomic databases
PIR 118224 108103 0.22 28 Sequence databases
PIRSF 98487 98472 0.18 29 Family and domain databases
PMAP-CutDB 1457 1457 <0.01 97 Other
PMMA-2DPAGE 52 52 <0.01 130 2D gel databases
PomBase 5016 4958 0.01 82 Organism-specific databases
PptaseDB 38 38 <0.01 131 Protein family/group databases
PRIDE 79833 79833 0.15 36 Proteomic databases
PRINTS 137353 120147 0.26 26 Family and domain databases
ProDom 29293 29114 0.05 52 Family and domain databases
ProMEX 513 513 <0.01 113 Proteomic databases
PROSITE 480044 303359 0.89 7 Family and domain databases
ProtClustDB 343244 343244 0.64 15 Phylogenomic databases
ProteinModelPortal 429127 429127 0.80 10 3D structure databases
PseudoCAP 1250 1241 <0.01 100 Organism-specific databases
Rat-heart-2DPAGE 28 28 <0.01 133 2D gel databases
Reactome 12221 7551 0.02 69 Enzyme and pathway databases
REBASE 402 402 <0.01 116 Protein family/group databases
RefSeq 521452 469836 0.97 5 Sequence databases
REPRODUCTION-2DPAGE 1256 1035 <0.01 99 2D gel databases
RGD 7724 7720 0.01 73 Organism-specific databases
SGD 6640 6635 0.01 76 Organism-specific databases
Siena-2DPAGE 103 103 <0.01 125 2D gel databases
SMART 167200 125162 0.31 25 Family and domain databases
SMR 218856 218856 0.41 23 3D structure databases
STRING 309313 309310 0.57 19 Protein-protein interaction databases
SUPFAM 330871 261981 0.61 16 Family and domain databases
SWISS-2DPAGE 1182 1181 <0.01 101 2D gel databases
TAIR 11588 11535 0.02 71 Organism-specific databases
TCDB 3647 3632 0.01 93 Protein family/group databases
TIGR 35 33 <0.01 132 Genome annotation databases
TIGRFAMs 289887 269320 0.54 21 Family and domain databases
TubercuList 1982 1946 <0.01 96 Organism-specific databases
UCD-2DPAGE 509 500 <0.01 114 2D gel databases
UCSC 51926 39791 0.10 44 Genome annotation databases
UniGene 98447 89672 0.18 30 Sequence databases
UniPathway 131304 119154 0.24 27 Enzyme and pathway databases
VectorBase 606 588 <0.01 112 Genome annotation databases
World-2DPAGE 919 908 <0.01 103 2D gel databases
WormBase 4798 3880 0.01 84 Organism-specific databases
Xenbase 4717 4712 0.01 85 Organism-specific databases
ZFIN 2775 2775 0.01 95 Organism-specific databases
Total number of cross-referenced databases: 134
6. AMINO ACID COMPOSITION
6.1 Composition in percent for the complete database
Ala (A) 8.25 Gln (Q) 3.93 Leu (L) 9.66 Ser (S) 6.56
Arg (R) 5.53 Glu (E) 6.75 Lys (K) 5.84 Thr (T) 5.34
Asn (N) 4.06 Gly (G) 7.08 Met (M) 2.42 Trp (W) 1.08
Asp (D) 5.45 His (H) 2.27 Phe (F) 3.86 Tyr (Y) 2.92
Cys (C) 1.37 Ile (I) 5.96 Pro (P) 4.70 Val (V) 6.87
Asx (B) 0.000 Glx (Z) 0.000 Xaa (X) 0.00
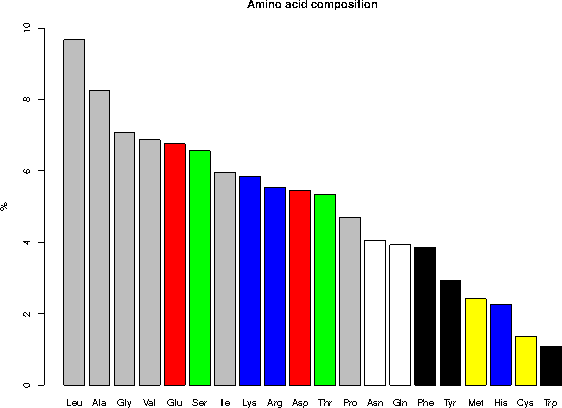 Legend: gray = aliphatic, red = acidic, green = small hydroxy,
blue = basic, black = aromatic, white = amide, yellow = sulfur
6.2 Classification of the amino acids by their frequency
Leu, Ala, Gly, Val, Glu, Ser, Ile, Lys, Arg, Asp, Thr, Pro, Asn, Gln,
Phe, Tyr, Met, His, Cys, Trp
7. MISCELLANEOUS STATISTICS
4460 entries are encoded on a mitochondrion, and 3687 are encoded on a plasmid.
12188 entries are encoded on a plastid,
of which 21 are encoded on apicoplasts,
11623 on chloroplasts,
51 on organellar chromatophores,
145 on cyanelles,
149 on non-photosynthetic plastids and
199 on unspecified types of plastid.
Number of entries with at least one sequence correction: 74058
Legend: gray = aliphatic, red = acidic, green = small hydroxy,
blue = basic, black = aromatic, white = amide, yellow = sulfur
6.2 Classification of the amino acids by their frequency
Leu, Ala, Gly, Val, Glu, Ser, Ile, Lys, Arg, Asp, Thr, Pro, Asn, Gln,
Phe, Tyr, Met, His, Cys, Trp
7. MISCELLANEOUS STATISTICS
4460 entries are encoded on a mitochondrion, and 3687 are encoded on a plasmid.
12188 entries are encoded on a plastid,
of which 21 are encoded on apicoplasts,
11623 on chloroplasts,
51 on organellar chromatophores,
145 on cyanelles,
149 on non-photosynthetic plastids and
199 on unspecified types of plastid.
Number of entries with at least one sequence correction: 74058
 2. TAXONOMIC ORIGIN
Total number of species represented in this release of UniProtKB/Swiss-Prot: 12922
The first twenty species represent 112553 sequences: 20.9 % of the total
number of entries.
2.1 Table of the frequency of occurrence of species
Species represented 1x: 5426
2x: 1882
3x: 981
4x: 639
5x: 466
6x: 381
7x: 284
8x: 217
9x: 199
10x: 123
11- 20x: 668
21- 50x: 403
51-100x: 212
>100x: 1041
2.2 Table of the most represented species
------ --------- --------------------------------------------
Number Frequency Species
------ --------- --------------------------------------------
1 20233 Homo sapiens (Human)
2 16566 Mus musculus (Mouse)
3 11571 Arabidopsis thaliana (Mouse-ear cress)
4 7815 Rattus norvegicus (Rat)
5 6621 Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
6 5965 Bos taurus (Bovine)
7 5089 Schizosaccharomyces pombe (strain 972 / ATCC 24843) (Fission yeast)
8 4431 Escherichia coli (strain K12)
9 4188 Bacillus subtilis (strain 168)
10 4126 Dictyostelium discoideum (Slime mold)
11 3364 Caenorhabditis elegans
12 3356 Xenopus laevis (African clawed frog)
13 3165 Drosophila melanogaster (Fruit fly)
14 2967 Oryza sativa subsp. japonica (Rice)
15 2861 Danio rerio (Zebrafish) (Brachydanio rerio)
16 2252 Gallus gallus (Chicken)
17 2218 Pongo abelii (Sumatran orangutan) (Pongo pygmaeus abelii)
18 2012 Escherichia coli O157:H7
19 1966 Mycobacterium tuberculosis
20 1787 Methanocaldococcus jannaschii
21 1770 Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)
22 1707 Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd)
23 1678 Shigella flexneri
24 1678 Xenopus tropicalis (Western clawed frog) (Silurana tropicalis)
25 1675 Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC)
26 1409 Sus scrofa (Pig)
27 1346 Salmonella typhi
28 1242 Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 / 1C / PRS 101 / LMG 12228)
29 1241 Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97)
30 1170 Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
31 1036 Synechocystis sp. (strain PCC 6803 / Kazusa)
32 1018 Yersinia pestis
33 1012 Archaeoglobus fulgidus
34 949 Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)
35 930 Salmonella paratyphi A (strain ATCC 9150 / SARB42)
36 926 Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056)
37 925 Staphylococcus aureus (strain N315)
38 923 Staphylococcus aureus (strain Mu50 / ATCC 700699)
39 909 Acanthamoeba polyphaga mimivirus (APMV)
40 904 Kluyveromyces lactis
41 899 Staphylococcus aureus (strain COL)
42 895 Staphylococcus aureus (strain MW2)
43 889 Staphylococcus aureus (strain MSSA476)
44 888 Escherichia coli O6:K15:H31 (strain 536 / UPEC)
45 888 Staphylococcus aureus (strain MRSA252)
46 886 Oryctolagus cuniculus (Rabbit)
47 882 Salmonella choleraesuis (strain SC-B67)
48 878 Shigella sonnei (strain Ss046)
49 869 Rhizobium meliloti (strain 1021) (Ensifer meliloti) (Sinorhizobium meliloti)
50 863 Yersinia pseudotuberculosis serotype I (strain IP32953)
51 861 Candida glabrata
52 841 Escherichia coli O9:H4 (strain HS)
53 834 Neurospora crassa
54 834 Escherichia coli O139:H28 (strain E24377A / ETEC)
55 829 Shigella boydii serotype 4 (strain Sb227)
56 825 Escherichia coli (strain UTI89 / UPEC)
57 821 Shigella dysenteriae serotype 1 (strain Sd197)
58 819 Escherichia coli (strain ATCC 8739 / DSM 1576 / Crooks)
59 803 Canis familiaris (Dog) (Canis lupus familiaris)
60 791 Escherichia coli (strain SMS-3-5 / SECEC)
61 787 Vibrio parahaemolyticus serotype O3:K6 (strain RIMD 2210633)
62 783 Erwinia carotovora subsp. atroseptica (strain SCRI 1043 / ATCC BAA-672)
63 782 Aquifex aeolicus (strain VF5)
64 775 Pasteurella multocida (strain Pm70)
65 773 Emericella nidulans
66 771 Escherichia coli (strain K12 / DH10B)
67 765 Escherichia coli O127:H6 (strain E2348/69 / EPEC)
68 765 Escherichia coli (strain K12 / MC4100 / BW2952)
69 764 Escherichia coli O17:K52:H18 (strain UMN026 / ExPEC)
70 762 Escherichia coli (strain 55989 / EAEC)
71 761 Escherichia coli O8 (strain IAI1)
72 760 Shigella flexneri serotype 5b (strain 8401)
73 759 Staphylococcus epidermidis (strain ATCC 35984 / RP62A)
74 758 Staphylococcus epidermidis (strain ATCC 12228)
75 756 Escherichia coli (strain SE11)
76 756 Escherichia coli O45:K1 (strain S88 / ExPEC)
77 754 Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145)
78 753 Escherichia coli O7:K1 (strain IAI39 / ExPEC)
79 748 Escherichia coli O157:H7 (strain EC4115 / EHEC)
80 744 Photorhabdus luminescens subsp. laumondii (strain TT01)
81 740 Staphylococcus aureus (strain NCTC 8325)
82 735 Bacillus halodurans
83 735 Yersinia enterocolitica serotype O:8 / biotype 1B (strain 8081)
84 734 Bacillus anthracis
85 732 Vibrio vulnificus (strain CMCP6)
86 731 Escherichia coli O81 (strain ED1a)
87 721 Salmonella enteritidis PT4 (strain P125109)
88 717 Vibrio vulnificus (strain YJ016)
89 716 Salmonella paratyphi B (strain ATCC BAA-1250 / SPB7)
90 715 Yersinia pestis bv. Antiqua (strain Nepal516)
91 714 Salmonella paratyphi A (strain AKU_12601)
92 713 Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578)
93 713 Enterobacter sp. (strain 638)
94 713 Salmonella agona (strain SL483)
95 713 Escherichia coli O1:K1 / APEC
96 713 Salmonella newport (strain SL254)
97 713 Yersinia pseudotuberculosis serotype O:1b (strain IP 31758)
98 712 Salmonella schwarzengrund (strain CVM19633)
99 711 Yersinia pestis bv. Antiqua (strain Antiqua)
100 711 Zea mays (Maize)
101 710 Salmonella heidelberg (strain SL476)
102 702 Salmonella dublin (strain CT_02021853)
103 701 Candida albicans (strain SC5314 / ATCC MYA-2876) (Yeast)
104 698 Shigella boydii serotype 18 (strain CDC 3083-94 / BS512)
105 696 Klebsiella pneumoniae (strain 342)
106 695 Escherichia fergusonii (strain ATCC 35469 / DSM 13698 / CDC 0568-73)
107 693 Pseudomonas putida (strain KT2440)
108 689 Nostoc sp. (strain PCC 7120 / UTEX 2576)
109 688 Pan troglodytes (Chimpanzee)
110 687 Mycoplasma pneumoniae (strain ATCC 29342 / M129)
111 683 Salmonella gallinarum (strain 287/91 / NCTC 13346)
112 678 Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
113 675 Pseudomonas syringae pv. tomato (strain DC3000)
114 670 Serratia proteamaculans (strain 568)
115 668 Mycobacterium leprae (strain TN)
116 667 Yersinia pestis (strain Pestoides F)
117 666 Staphylococcus aureus (strain USA300)
118 658 Rhizobium sp. (strain NGR234)
119 654 Bradyrhizobium japonicum (strain USDA 110)
120 653 Debaryomyces hansenii
121 652 Bacillus cereus (strain ATCC 14579 / DSM 31)
122 650 Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100)
123 645 Escherichia coli
124 643 Staphylococcus aureus (strain bovine RF122 / ET3-1)
125 642 Salmonella arizonae (strain ATCC BAA-731 / CDC346-86 / RSK2980)
126 642 Yarrowia lipolytica (strain CLIB 122 / E 150) (Yeast) (Candida lipolytica)
127 638 Yersinia pseudotuberculosis serotype O:3 (strain YPIII)
128 634 Shewanella oneidensis (strain MR-1)
129 634 Yersinia pseudotuberculosis serotype IB (strain PB1/+)
130 632 Agrobacterium tumefaciens (strain C58 / ATCC 33970)
131 622 Cronobacter sakazakii (strain ATCC BAA-894) (Enterobacter sakazakii)
132 616 Treponema pallidum (strain Nichols)
133 615 Oryza sativa subsp. indica (Rice)
134 613 Methanothermobacter thermautotrophicus
135 612 Staphylococcus haemolyticus (strain JCSC1435)
136 606 Rhizobium loti (strain MAFF303099) (Mesorhizobium loti)
137 605 Xanthomonas campestris pv. campestris (strain ATCC 33913 / NCPPB 528 / LMG 568)
138 602 Ralstonia solanacearum (strain GMI1000) (Pseudomonas solanacearum)
139 602 Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e)
140 602 Photobacterium profundum (Photobacterium sp. (strain SS9))
141 602 Staphylococcus saprophyticus subsp. saprophyticus
142 601 Salmonella paratyphi C (strain RKS4594)
143 600 Yersinia pestis bv. Antiqua (strain Angola)
144 590 Bacillus cereus (strain ATCC 10987)
145 590 Listeria innocua serovar 6a (strain CLIP 11262)
146 589 Pectobacterium carotovorum subsp. carotovorum (strain PC1)
147 586 Rickettsia prowazekii (strain Madrid E)
148 580 Helicobacter pylori (strain ATCC 700392 / 26695) (Campylobacter pylori)
149 578 Neisseria meningitidis serogroup B (strain MC58)
150 576 Brucella suis biovar 1 (strain 1330)
151 572 Brucella melitensis biotype 1 (strain 16M / ATCC 23456 / NCTC 10094)
152 572 Buchnera aphidicola subsp. Acyrthosiphon pisum (strain APS)
153 568 Caenorhabditis briggsae
154 567 Bacillus thuringiensis subsp. konkukian (strain 97-27)
155 566 Pseudomonas syringae pv. syringae (strain B728a)
156 565 Helicobacter pylori (strain J99) (Campylobacter pylori J99)
157 564 Vibrio fischeri (strain ATCC 700601 / ES114)
158 564 Caulobacter crescentus (strain ATCC 19089 / CB15)
159 564 Pseudomonas aeruginosa (strain UCBPP-PA14)
160 562 Bacillus licheniformis (strain DSM 13 / ATCC 14580)
161 562 Buchnera aphidicola subsp. Schizaphis graminum (strain Sg)
162 561 Bacillus cereus (strain ZK / E33L)
163 556 Xanthomonas axonopodis pv. citri (strain 306)
164 556 Clostridium acetobutylicum
165 552 Oceanobacillus iheyensis (strain DSM 14371 / JCM 11309 / KCTC 3954 / HTE831)
166 552 Neisseria meningitidis serogroup A / serotype 4A (strain Z2491)
167 552 Pseudomonas fluorescens (strain Pf0-1)
168 546 Pseudomonas fluorescens (strain Pf-5 / ATCC BAA-477)
169 545 Pseudomonas syringae pv. phaseolicola (strain 1448A / Race 6)
170 533 Lactococcus lactis subsp. lactis (strain IL1403) (Streptococcus lactis)
171 531 Erwinia tasmaniensis (strain DSM 17950 / Et1/99)
172 529 Sodalis glossinidius (strain morsitans)
173 529 Listeria monocytogenes serotype 4b (strain F2365)
174 527 Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099)
175 522 Bordetella bronchiseptica (strain ATCC BAA-588 / NCTC 13252 / RB50)
176 522 Xylella fastidiosa (strain 9a5c)
177 515 Chromobacterium violaceum
178 515 Bordetella pertussis (strain Tohama I / ATCC BAA-589 / NCTC 13251)
179 512 Xylella fastidiosa (strain Temecula1 / ATCC 700964)
180 511 Pseudomonas aeruginosa (strain PA7)
181 511 Vibrio cholerae serotype O1 (strain ATCC 39541 / Ogawa 395 / O395)
182 510 Haemophilus ducreyi (strain 35000HP / ATCC 700724)
183 508 Bordetella parapertussis (strain 12822 / ATCC BAA-587 / NCTC 13253)
184 507 Buchnera aphidicola subsp. Baizongia pistaciae (strain Bp)
185 507 Geobacillus kaustophilus (strain HTA426)
186 506 Staphylococcus aureus (strain Newman)
187 502 Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4)
188 501 Deinococcus radiodurans
189 500 Pseudomonas entomophila (strain L48)
190 499 Corynebacterium glutamicum
191 499 Brucella abortus biovar 1 (strain 9-941)
192 497 Streptomyces avermitilis
193 497 Rickettsia conorii (strain ATCC VR-613 / Malish 7)
194 496 Bacillus clausii (strain KSM-K16)
195 495 Burkholderia pseudomallei (strain K96243)
196 495 Haemophilus influenzae (strain 86-028NP)
197 494 Proteus mirabilis (strain HI4320)
198 492 Bacillus amyloliquefaciens (strain FZB42)
199 491 Xanthomonas campestris pv. campestris (strain 8004)
200 490 Vibrio harveyi (strain ATCC BAA-1116 / BB120)
201 489 Methanosarcina acetivorans (strain ATCC 35395 / DSM 2834 / JCM 12185 / C2A)
202 487 Shewanella sp. (strain MR-7)
203 486 Mannheimia succiniciproducens (strain MBEL55E)
204 484 Pseudomonas aeruginosa (strain LESB58)
205 484 Staphylococcus aureus (strain Mu3 / ATCC 700698)
206 484 Shewanella sp. (strain MR-4)
207 483 Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195)
208 481 Thermosynechococcus elongatus (strain BP-1)
209 480 Acinetobacter sp. (strain ADP1)
210 479 Pyrococcus horikoshii
211 478 Synechococcus elongatus (strain PCC 7942) (Anacystis nidulans R2)
212 475 Pseudomonas putida (strain F1 / ATCC 700007)
213 474 Burkholderia sp. (strain 383) (Burkholderia cepacia
214 474 Brucella abortus (strain 2308)
215 473 Aspergillus oryzae (strain ATCC 42149 / RIB 40) (Yellow koji mold)
216 472 Streptococcus pneumoniae (strain ATCC BAA-255 / R6)
217 469 Methanosarcina mazei
218 467 Halobacterium salinarum (strain ATCC 700922 / JCM 11081 / NRC-1)
219 467 Clostridium perfringens (strain 13 / Type A)
220 466 Pyrococcus abyssi (strain GE5 / Orsay)
221 466 Xanthomonas campestris pv. vesicatoria (strain 85-10)
222 466 Shewanella frigidimarina (strain NCIMB 400)
223 466 Pseudomonas putida (strain GB-1)
224 466 Cupriavidus necator (strain ATCC 17699 / H16 / DSM 428 / Stanier 337)
225 464 Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / NCIB 9240)
226 463 Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1)
227 463 Shewanella sp. (strain ANA-3)
228 462 Burkholderia mallei (strain ATCC 23344)
229 462 Anabaena variabilis (strain ATCC 29413 / PCC 7937)
230 462 Rhodopseudomonas palustris (strain ATCC BAA-98 / CGA009)
231 461 Lactobacillus plantarum (strain ATCC BAA-793 / NCIMB 8826 / WCFS1)
232 459 Cupriavidus pinatubonensis (strain JMP134 / LMG 1197) (Alcaligenes eutrophus)
233 458 Enterococcus faecalis (strain ATCC 700802 / V583)
234 455 Staphylococcus aureus (strain JH1)
235 455 Rhodobacter sphaeroides (strain ATCC 17023 / 2.4.1 / NCIB 8253 / DSM 158)
236 454 Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath)
237 454 Xanthomonas oryzae pv. oryzae (strain MAFF 311018)
238 453 Ovis aries (Sheep)
239 453 Pseudomonas putida (strain W619)
240 453 Rickettsia felis (strain ATCC VR-1525 / URRWXCal2) (Rickettsia azadi)
241 452 Shewanella baltica (strain OS185)
242 451 Aeromonas salmonicida (strain A449)
243 449 Thermoanaerobacter tengcongensis
244 449 Staphylococcus aureus (strain JH9)
245 449 Hahella chejuensis (strain KCTC 2396)
246 448 Nicotiana tabacum (Common tobacco)
247 448 Mycobacterium paratuberculosis (strain ATCC BAA-968 / K-10)
248 448 Streptococcus mutans serotype c (strain ATCC 700610 / UA159)
249 447 Vibrio fischeri (strain MJ11)
250 446 Sulfolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2)
2.3 Taxonomic distribution of the sequences
2. TAXONOMIC ORIGIN
Total number of species represented in this release of UniProtKB/Swiss-Prot: 12922
The first twenty species represent 112553 sequences: 20.9 % of the total
number of entries.
2.1 Table of the frequency of occurrence of species
Species represented 1x: 5426
2x: 1882
3x: 981
4x: 639
5x: 466
6x: 381
7x: 284
8x: 217
9x: 199
10x: 123
11- 20x: 668
21- 50x: 403
51-100x: 212
>100x: 1041
2.2 Table of the most represented species
------ --------- --------------------------------------------
Number Frequency Species
------ --------- --------------------------------------------
1 20233 Homo sapiens (Human)
2 16566 Mus musculus (Mouse)
3 11571 Arabidopsis thaliana (Mouse-ear cress)
4 7815 Rattus norvegicus (Rat)
5 6621 Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
6 5965 Bos taurus (Bovine)
7 5089 Schizosaccharomyces pombe (strain 972 / ATCC 24843) (Fission yeast)
8 4431 Escherichia coli (strain K12)
9 4188 Bacillus subtilis (strain 168)
10 4126 Dictyostelium discoideum (Slime mold)
11 3364 Caenorhabditis elegans
12 3356 Xenopus laevis (African clawed frog)
13 3165 Drosophila melanogaster (Fruit fly)
14 2967 Oryza sativa subsp. japonica (Rice)
15 2861 Danio rerio (Zebrafish) (Brachydanio rerio)
16 2252 Gallus gallus (Chicken)
17 2218 Pongo abelii (Sumatran orangutan) (Pongo pygmaeus abelii)
18 2012 Escherichia coli O157:H7
19 1966 Mycobacterium tuberculosis
20 1787 Methanocaldococcus jannaschii
21 1770 Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)
22 1707 Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd)
23 1678 Shigella flexneri
24 1678 Xenopus tropicalis (Western clawed frog) (Silurana tropicalis)
25 1675 Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC)
26 1409 Sus scrofa (Pig)
27 1346 Salmonella typhi
28 1242 Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 / 1C / PRS 101 / LMG 12228)
29 1241 Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97)
30 1170 Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
31 1036 Synechocystis sp. (strain PCC 6803 / Kazusa)
32 1018 Yersinia pestis
33 1012 Archaeoglobus fulgidus
34 949 Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)
35 930 Salmonella paratyphi A (strain ATCC 9150 / SARB42)
36 926 Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056)
37 925 Staphylococcus aureus (strain N315)
38 923 Staphylococcus aureus (strain Mu50 / ATCC 700699)
39 909 Acanthamoeba polyphaga mimivirus (APMV)
40 904 Kluyveromyces lactis
41 899 Staphylococcus aureus (strain COL)
42 895 Staphylococcus aureus (strain MW2)
43 889 Staphylococcus aureus (strain MSSA476)
44 888 Escherichia coli O6:K15:H31 (strain 536 / UPEC)
45 888 Staphylococcus aureus (strain MRSA252)
46 886 Oryctolagus cuniculus (Rabbit)
47 882 Salmonella choleraesuis (strain SC-B67)
48 878 Shigella sonnei (strain Ss046)
49 869 Rhizobium meliloti (strain 1021) (Ensifer meliloti) (Sinorhizobium meliloti)
50 863 Yersinia pseudotuberculosis serotype I (strain IP32953)
51 861 Candida glabrata
52 841 Escherichia coli O9:H4 (strain HS)
53 834 Neurospora crassa
54 834 Escherichia coli O139:H28 (strain E24377A / ETEC)
55 829 Shigella boydii serotype 4 (strain Sb227)
56 825 Escherichia coli (strain UTI89 / UPEC)
57 821 Shigella dysenteriae serotype 1 (strain Sd197)
58 819 Escherichia coli (strain ATCC 8739 / DSM 1576 / Crooks)
59 803 Canis familiaris (Dog) (Canis lupus familiaris)
60 791 Escherichia coli (strain SMS-3-5 / SECEC)
61 787 Vibrio parahaemolyticus serotype O3:K6 (strain RIMD 2210633)
62 783 Erwinia carotovora subsp. atroseptica (strain SCRI 1043 / ATCC BAA-672)
63 782 Aquifex aeolicus (strain VF5)
64 775 Pasteurella multocida (strain Pm70)
65 773 Emericella nidulans
66 771 Escherichia coli (strain K12 / DH10B)
67 765 Escherichia coli O127:H6 (strain E2348/69 / EPEC)
68 765 Escherichia coli (strain K12 / MC4100 / BW2952)
69 764 Escherichia coli O17:K52:H18 (strain UMN026 / ExPEC)
70 762 Escherichia coli (strain 55989 / EAEC)
71 761 Escherichia coli O8 (strain IAI1)
72 760 Shigella flexneri serotype 5b (strain 8401)
73 759 Staphylococcus epidermidis (strain ATCC 35984 / RP62A)
74 758 Staphylococcus epidermidis (strain ATCC 12228)
75 756 Escherichia coli (strain SE11)
76 756 Escherichia coli O45:K1 (strain S88 / ExPEC)
77 754 Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145)
78 753 Escherichia coli O7:K1 (strain IAI39 / ExPEC)
79 748 Escherichia coli O157:H7 (strain EC4115 / EHEC)
80 744 Photorhabdus luminescens subsp. laumondii (strain TT01)
81 740 Staphylococcus aureus (strain NCTC 8325)
82 735 Bacillus halodurans
83 735 Yersinia enterocolitica serotype O:8 / biotype 1B (strain 8081)
84 734 Bacillus anthracis
85 732 Vibrio vulnificus (strain CMCP6)
86 731 Escherichia coli O81 (strain ED1a)
87 721 Salmonella enteritidis PT4 (strain P125109)
88 717 Vibrio vulnificus (strain YJ016)
89 716 Salmonella paratyphi B (strain ATCC BAA-1250 / SPB7)
90 715 Yersinia pestis bv. Antiqua (strain Nepal516)
91 714 Salmonella paratyphi A (strain AKU_12601)
92 713 Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578)
93 713 Enterobacter sp. (strain 638)
94 713 Salmonella agona (strain SL483)
95 713 Escherichia coli O1:K1 / APEC
96 713 Salmonella newport (strain SL254)
97 713 Yersinia pseudotuberculosis serotype O:1b (strain IP 31758)
98 712 Salmonella schwarzengrund (strain CVM19633)
99 711 Yersinia pestis bv. Antiqua (strain Antiqua)
100 711 Zea mays (Maize)
101 710 Salmonella heidelberg (strain SL476)
102 702 Salmonella dublin (strain CT_02021853)
103 701 Candida albicans (strain SC5314 / ATCC MYA-2876) (Yeast)
104 698 Shigella boydii serotype 18 (strain CDC 3083-94 / BS512)
105 696 Klebsiella pneumoniae (strain 342)
106 695 Escherichia fergusonii (strain ATCC 35469 / DSM 13698 / CDC 0568-73)
107 693 Pseudomonas putida (strain KT2440)
108 689 Nostoc sp. (strain PCC 7120 / UTEX 2576)
109 688 Pan troglodytes (Chimpanzee)
110 687 Mycoplasma pneumoniae (strain ATCC 29342 / M129)
111 683 Salmonella gallinarum (strain 287/91 / NCTC 13346)
112 678 Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
113 675 Pseudomonas syringae pv. tomato (strain DC3000)
114 670 Serratia proteamaculans (strain 568)
115 668 Mycobacterium leprae (strain TN)
116 667 Yersinia pestis (strain Pestoides F)
117 666 Staphylococcus aureus (strain USA300)
118 658 Rhizobium sp. (strain NGR234)
119 654 Bradyrhizobium japonicum (strain USDA 110)
120 653 Debaryomyces hansenii
121 652 Bacillus cereus (strain ATCC 14579 / DSM 31)
122 650 Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100)
123 645 Escherichia coli
124 643 Staphylococcus aureus (strain bovine RF122 / ET3-1)
125 642 Salmonella arizonae (strain ATCC BAA-731 / CDC346-86 / RSK2980)
126 642 Yarrowia lipolytica (strain CLIB 122 / E 150) (Yeast) (Candida lipolytica)
127 638 Yersinia pseudotuberculosis serotype O:3 (strain YPIII)
128 634 Shewanella oneidensis (strain MR-1)
129 634 Yersinia pseudotuberculosis serotype IB (strain PB1/+)
130 632 Agrobacterium tumefaciens (strain C58 / ATCC 33970)
131 622 Cronobacter sakazakii (strain ATCC BAA-894) (Enterobacter sakazakii)
132 616 Treponema pallidum (strain Nichols)
133 615 Oryza sativa subsp. indica (Rice)
134 613 Methanothermobacter thermautotrophicus
135 612 Staphylococcus haemolyticus (strain JCSC1435)
136 606 Rhizobium loti (strain MAFF303099) (Mesorhizobium loti)
137 605 Xanthomonas campestris pv. campestris (strain ATCC 33913 / NCPPB 528 / LMG 568)
138 602 Ralstonia solanacearum (strain GMI1000) (Pseudomonas solanacearum)
139 602 Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e)
140 602 Photobacterium profundum (Photobacterium sp. (strain SS9))
141 602 Staphylococcus saprophyticus subsp. saprophyticus
142 601 Salmonella paratyphi C (strain RKS4594)
143 600 Yersinia pestis bv. Antiqua (strain Angola)
144 590 Bacillus cereus (strain ATCC 10987)
145 590 Listeria innocua serovar 6a (strain CLIP 11262)
146 589 Pectobacterium carotovorum subsp. carotovorum (strain PC1)
147 586 Rickettsia prowazekii (strain Madrid E)
148 580 Helicobacter pylori (strain ATCC 700392 / 26695) (Campylobacter pylori)
149 578 Neisseria meningitidis serogroup B (strain MC58)
150 576 Brucella suis biovar 1 (strain 1330)
151 572 Brucella melitensis biotype 1 (strain 16M / ATCC 23456 / NCTC 10094)
152 572 Buchnera aphidicola subsp. Acyrthosiphon pisum (strain APS)
153 568 Caenorhabditis briggsae
154 567 Bacillus thuringiensis subsp. konkukian (strain 97-27)
155 566 Pseudomonas syringae pv. syringae (strain B728a)
156 565 Helicobacter pylori (strain J99) (Campylobacter pylori J99)
157 564 Vibrio fischeri (strain ATCC 700601 / ES114)
158 564 Caulobacter crescentus (strain ATCC 19089 / CB15)
159 564 Pseudomonas aeruginosa (strain UCBPP-PA14)
160 562 Bacillus licheniformis (strain DSM 13 / ATCC 14580)
161 562 Buchnera aphidicola subsp. Schizaphis graminum (strain Sg)
162 561 Bacillus cereus (strain ZK / E33L)
163 556 Xanthomonas axonopodis pv. citri (strain 306)
164 556 Clostridium acetobutylicum
165 552 Oceanobacillus iheyensis (strain DSM 14371 / JCM 11309 / KCTC 3954 / HTE831)
166 552 Neisseria meningitidis serogroup A / serotype 4A (strain Z2491)
167 552 Pseudomonas fluorescens (strain Pf0-1)
168 546 Pseudomonas fluorescens (strain Pf-5 / ATCC BAA-477)
169 545 Pseudomonas syringae pv. phaseolicola (strain 1448A / Race 6)
170 533 Lactococcus lactis subsp. lactis (strain IL1403) (Streptococcus lactis)
171 531 Erwinia tasmaniensis (strain DSM 17950 / Et1/99)
172 529 Sodalis glossinidius (strain morsitans)
173 529 Listeria monocytogenes serotype 4b (strain F2365)
174 527 Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099)
175 522 Bordetella bronchiseptica (strain ATCC BAA-588 / NCTC 13252 / RB50)
176 522 Xylella fastidiosa (strain 9a5c)
177 515 Chromobacterium violaceum
178 515 Bordetella pertussis (strain Tohama I / ATCC BAA-589 / NCTC 13251)
179 512 Xylella fastidiosa (strain Temecula1 / ATCC 700964)
180 511 Pseudomonas aeruginosa (strain PA7)
181 511 Vibrio cholerae serotype O1 (strain ATCC 39541 / Ogawa 395 / O395)
182 510 Haemophilus ducreyi (strain 35000HP / ATCC 700724)
183 508 Bordetella parapertussis (strain 12822 / ATCC BAA-587 / NCTC 13253)
184 507 Buchnera aphidicola subsp. Baizongia pistaciae (strain Bp)
185 507 Geobacillus kaustophilus (strain HTA426)
186 506 Staphylococcus aureus (strain Newman)
187 502 Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4)
188 501 Deinococcus radiodurans
189 500 Pseudomonas entomophila (strain L48)
190 499 Corynebacterium glutamicum
191 499 Brucella abortus biovar 1 (strain 9-941)
192 497 Streptomyces avermitilis
193 497 Rickettsia conorii (strain ATCC VR-613 / Malish 7)
194 496 Bacillus clausii (strain KSM-K16)
195 495 Burkholderia pseudomallei (strain K96243)
196 495 Haemophilus influenzae (strain 86-028NP)
197 494 Proteus mirabilis (strain HI4320)
198 492 Bacillus amyloliquefaciens (strain FZB42)
199 491 Xanthomonas campestris pv. campestris (strain 8004)
200 490 Vibrio harveyi (strain ATCC BAA-1116 / BB120)
201 489 Methanosarcina acetivorans (strain ATCC 35395 / DSM 2834 / JCM 12185 / C2A)
202 487 Shewanella sp. (strain MR-7)
203 486 Mannheimia succiniciproducens (strain MBEL55E)
204 484 Pseudomonas aeruginosa (strain LESB58)
205 484 Staphylococcus aureus (strain Mu3 / ATCC 700698)
206 484 Shewanella sp. (strain MR-4)
207 483 Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195)
208 481 Thermosynechococcus elongatus (strain BP-1)
209 480 Acinetobacter sp. (strain ADP1)
210 479 Pyrococcus horikoshii
211 478 Synechococcus elongatus (strain PCC 7942) (Anacystis nidulans R2)
212 475 Pseudomonas putida (strain F1 / ATCC 700007)
213 474 Burkholderia sp. (strain 383) (Burkholderia cepacia
214 474 Brucella abortus (strain 2308)
215 473 Aspergillus oryzae (strain ATCC 42149 / RIB 40) (Yellow koji mold)
216 472 Streptococcus pneumoniae (strain ATCC BAA-255 / R6)
217 469 Methanosarcina mazei
218 467 Halobacterium salinarum (strain ATCC 700922 / JCM 11081 / NRC-1)
219 467 Clostridium perfringens (strain 13 / Type A)
220 466 Pyrococcus abyssi (strain GE5 / Orsay)
221 466 Xanthomonas campestris pv. vesicatoria (strain 85-10)
222 466 Shewanella frigidimarina (strain NCIMB 400)
223 466 Pseudomonas putida (strain GB-1)
224 466 Cupriavidus necator (strain ATCC 17699 / H16 / DSM 428 / Stanier 337)
225 464 Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / NCIB 9240)
226 463 Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1)
227 463 Shewanella sp. (strain ANA-3)
228 462 Burkholderia mallei (strain ATCC 23344)
229 462 Anabaena variabilis (strain ATCC 29413 / PCC 7937)
230 462 Rhodopseudomonas palustris (strain ATCC BAA-98 / CGA009)
231 461 Lactobacillus plantarum (strain ATCC BAA-793 / NCIMB 8826 / WCFS1)
232 459 Cupriavidus pinatubonensis (strain JMP134 / LMG 1197) (Alcaligenes eutrophus)
233 458 Enterococcus faecalis (strain ATCC 700802 / V583)
234 455 Staphylococcus aureus (strain JH1)
235 455 Rhodobacter sphaeroides (strain ATCC 17023 / 2.4.1 / NCIB 8253 / DSM 158)
236 454 Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath)
237 454 Xanthomonas oryzae pv. oryzae (strain MAFF 311018)
238 453 Ovis aries (Sheep)
239 453 Pseudomonas putida (strain W619)
240 453 Rickettsia felis (strain ATCC VR-1525 / URRWXCal2) (Rickettsia azadi)
241 452 Shewanella baltica (strain OS185)
242 451 Aeromonas salmonicida (strain A449)
243 449 Thermoanaerobacter tengcongensis
244 449 Staphylococcus aureus (strain JH9)
245 449 Hahella chejuensis (strain KCTC 2396)
246 448 Nicotiana tabacum (Common tobacco)
247 448 Mycobacterium paratuberculosis (strain ATCC BAA-968 / K-10)
248 448 Streptococcus mutans serotype c (strain ATCC 700610 / UA159)
249 447 Vibrio fischeri (strain MJ11)
250 446 Sulfolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2)
2.3 Taxonomic distribution of the sequences
 Kingdom sequences (% of the database)
Archaea 18953 ( 4%)
Bacteria 328550 ( 61%)
Eukaryota 174462 ( 32%)
Viruses 16294 ( 3%)
Within Eukaryota:
Kingdom sequences (% of the database)
Archaea 18953 ( 4%)
Bacteria 328550 ( 61%)
Eukaryota 174462 ( 32%)
Viruses 16294 ( 3%)
Within Eukaryota:
 Category sequences (% of Eukaryota) (% of the complete database)
Human 20234 ( 12%) ( 4%)
Other Mammalia 45763 ( 26%) ( 9%)
Other Vertebrata 17270 ( 10%) ( 3%)
Viridiplantae 33169 ( 19%) ( 6%)
Fungi 30780 ( 18%) ( 6%)
Insecta 8425 ( 5%) ( 2%)
Nematoda 4238 ( 2%) ( 1%)
Other 14583 ( 8%) ( 3%)
3. SEQUENCE SIZE
Repartition of the sequences by size (excluding fragments)
From To Number From To Number
1- 50 8735 1001-1100 3733
51- 100 41281 1101-1200 2586
101- 150 57513 1201-1300 2009
151- 200 57638 1301-1400 1870
201- 250 56409 1401-1500 1502
251- 300 49836 1501-1600 725
301- 350 49996 1601-1700 562
351- 400 43196 1701-1800 459
401- 450 35392 1801-1900 423
451- 500 28489 1901-2000 349
501- 550 20245 2001-2100 212
551- 600 14523 2101-2200 283
601- 650 12217 2201-2300 292
651- 700 8818 2301-2400 172
701- 750 7262 2401-2500 136
751- 800 5159 >2500 1081
801- 850 4523
851- 900 5009
901- 950 3851
951-1000 2733
Category sequences (% of Eukaryota) (% of the complete database)
Human 20234 ( 12%) ( 4%)
Other Mammalia 45763 ( 26%) ( 9%)
Other Vertebrata 17270 ( 10%) ( 3%)
Viridiplantae 33169 ( 19%) ( 6%)
Fungi 30780 ( 18%) ( 6%)
Insecta 8425 ( 5%) ( 2%)
Nematoda 4238 ( 2%) ( 1%)
Other 14583 ( 8%) ( 3%)
3. SEQUENCE SIZE
Repartition of the sequences by size (excluding fragments)
From To Number From To Number
1- 50 8735 1001-1100 3733
51- 100 41281 1101-1200 2586
101- 150 57513 1201-1300 2009
151- 200 57638 1301-1400 1870
201- 250 56409 1401-1500 1502
251- 300 49836 1501-1600 725
301- 350 49996 1601-1700 562
351- 400 43196 1701-1800 459
401- 450 35392 1801-1900 423
451- 500 28489 1901-2000 349
501- 550 20245 2001-2100 212
551- 600 14523 2101-2200 283
601- 650 12217 2201-2300 292
651- 700 8818 2301-2400 172
701- 750 7262 2401-2500 136
751- 800 5159 >2500 1081
801- 850 4523
851- 900 5009
901- 950 3851
951-1000 2733
 The average sequence length in UniProtKB/Swiss-Prot is 355 amino acids.
The shortest sequence is GWA_SEPOF (P83570): 2 amino acids.
The longest sequence is TITIN_MOUSE (A2ASS6): 35213 amino acids.
4. JOURNAL CITATIONS
Note: the following citation statistics reflect the number of distinct
journal citations.
Total number of journals cited in this release of UniProtKB/Swiss-Prot: 2280
4.1 Table of the frequency of journal citations
Journals cited 1x: 751
2x: 290
3x: 152
4x: 118
5x: 84
6x: 69
7x: 57
8x: 45
9x: 32
10x: 29
11- 20x: 182
21- 50x: 190
51-100x: 100
>100x: 181
4.2 List of the most cited journals in UniProtKB/Swiss-Prot
Nb Citations Journal name
-- --------- -------------------------------------------------------------
1 20577 Journal of Biological Chemistry
2 9326 Proceedings of the National Academy of Sciences of the U.S.A.
3 5604 Journal of Bacteriology
4 4973 Biochemical and Biophysical Research Communications
5 4585 Gene
6 4500 Nucleic Acids Research
7 4342 Biochemistry
8 4299 FEBS Letters
9 4155 The EMBO Journal
10 3868 Molecular and Cellular Biology
11 3636 Nature
12 3479 Journal of Molecular Biology
13 3226 European Journal of Biochemistry
14 3180 Biochimica et Biophysica Acta
15 2976 Cell
16 2503 Genomics
17 2463 Journal of Virology
18 2428 Biochemical Journal
19 2399 Science
20 1989 Molecular Microbiology
21 1822 Journal of Cell Biology
22 1769 Plant Physiology
23 1614 Plant Molecular Biology
24 1559 The American Journal of Human Genetics
25 1553 Genes and Development
26 1513 Virology
27 1476 Nature Genetics
28 1433 Human Molecular Genetics
29 1405 Oncogene
30 1331 Molecular and General Genetics
31 1304 Development
32 1290 Human Mutation
33 1255 Molecular Biology of the Cell
34 1225 The Plant Cell
35 1224 Journal of Biochemistry
36 1158 Journal of Immunology
37 1108 Molecular Cell
38 1095 The Plant Journal
39 1090 Genetics
40 1046 Structure
41 1013 Journal of General Virology
42 957 Blood
43 929 Infection and Immunity
44 918 Archives of Biochemistry and Biophysics
45 909 Journal of Cell Science
46 831 Microbiology
47 816 Developmental Biology
48 794 Cancer Research
49 783 Yeast
50 772 Current Biology
51 715 FEMS Microbiology Letters
52 649 Acta Crystallographica, Section D
53 636 Protein Science
54 629 Journal of Neuroscience
55 623 Applied and Environmental Microbiology
56 623 Toxicon
57 622 Human Genetics
58 618 Nature Structural Biology
59 616 Mechanisms of Development
60 579 Neuron
61 565 Journal of Clinical Investigation
62 537 Current Genetics
63 532 American Journal of Physiology
64 522 The Journal of Experimental Medicine
65 483 Proteins
66 481 Molecular Endocrinology
67 479 Mammalian Genome
68 457 Journal of Neurochemistry
69 457 PLoS ONE
70 456 Immunogenetics
71 441 The Journal of Clinical Endocrinology and Metabolism
72 441 Plant and Cell Physiology
73 433 Bioscience, Biotechnology, and Biochemistry
74 430 Endocrinology
75 428 Molecular and Biochemical Parasitology
76 423 Nature Cell Biology
77 400 Journal of Medical Genetics
78 395 Journal of Molecular Evolution
79 377 DNA and Cell Biology
80 375 Molecular Biology and Evolution
81 367 Experimental Cell Research
82 364 DNA Sequence
83 342 Peptides
84 326 Brain Research. Molecular Brain Research
85 324 Tissue Antigens
86 318 Comparative Biochemistry and Physiology
87 311 Developmental Cell
88 310 RNA
89 310 Antimicrobial Agents and Chemotherapy
90 303 Journal of Investigative Dermatology
91 302 Molecular Pharmacology
92 294 Biological Chemistry Hoppe-Seyler
93 293 Nature Structural and Molecular Biology
94 290 Planta
95 288 The FEBS Journal
96 280 Biology of Reproduction
97 279 Cytogenetics and Cell Genetics
98 277 Neurology
99 266 Developmental Dynamics
100 263 Genome Research
101 262 Virus Research
102 254 Journal of General Microbiology
103 248 EMBO Reports
104 245 Immunity
105 243 Molecular Plant-Microbe Interactions
106 243 Biochimie
107 235 Eukaryotic Cell
108 233 The New England Journal of Medicine
109 232 The FASEB Journal
110 232 Genes to Cells
111 227 European Journal of Immunology
112 222 Annals of Neurology
113 219 European Journal of Human Genetics
114 218 Hoppe-Seyler's Zeitschrift fur Physiologische Chemie
115 212 DNA Research
116 209 Journal of Human Genetics
117 197 Investigative Ophthalmology and Visual Science
118 196 Archives of Microbiology
119 195 Journal of the American Chemical Society
120 188 Nature Immunology
121 188 Archives of Virology
122 184 Journal of Cellular Biochemistry
123 183 Molecular and Cellular Endocrinology
124 183 American Journal of Medical Genetics. Part A
125 182 BMC Genomics
126 180 Molecular Immunology
127 177 Glycobiology
128 176 Clinical Genetics
129 172 Insect Biochemistry and Molecular Biology
130 171 Diabetes
131 168 American Journal of Medical Genetics
132 167 Journal of Medicinal Chemistry
133 167 Molecular Phylogenetics and Evolution
134 165 Acta Crystallographica, Section F
135 165 Journal of Experimental Botany
136 163 Circulation Research
137 159 International Journal of Cancer
138 159 DNA
139 158 Molecular Reproduction and Development
140 155 Hemoglobin
141 154 Bioorganicheskaia Khimiia
142 154 Molecular Genetics and Metabolism
143 150 PLoS Genetics
144 149 Phytochemistry
145 149 Molecular and Cellular Neuroscience
146 149 Biological Chemistry
147 148 Molecular Genetics and Genomics
148 145 Journal of Lipid Research
149 145 Protein Expression and Purification
150 144 Traffic
5. STATISTICS FOR SOME LINE TYPES
The following table summarizes the total number of some UniProtKB/Swiss-Prot lines,
as well as the number of entries with at least one such line, and the
frequency of the lines.
Total Number of Average
Line type / subtype number entries per entry
------------------------------------ -------- --------- ---------
References (RL) 1028777 1.91
Journal 822062 413904 1.53 1
Submitted to EMBL/GenBank/DDBJ 197677 177199 0.37 2
Submitted to other databases 6851 6388 0.01 3
Book citation 717 703 <0.01 4
Plant Gene Register 578 566 <0.01 5
Thesis 413 410 <0.01 6
Unpublished observations 283 279 <0.01 7
Patent 190 187 <0.01 8
Worm Breeder's Gazette 6 6 <0.01 9
Total number of distinct authors cited in UniProtKB/Swiss-Prot: 328522
Total Number of Average
Line type / subtype number entries per entry Rank
------------------------------------ -------- --------- --------- ----
Comments (CC) 2400283 4.46
ALLERGEN 520 520 <0.01 26
ALTERNATIVE PRODUCTS 21332 21332 0.04 13
BIOPHYSICOCHEMICAL PROPERTIES 4624 4624 0.01 23
BIOTECHNOLOGY 332 330 <0.01 28
CATALYTIC ACTIVITY 242353 218947 0.45 4
CAUTION 8087 7921 0.02 19
COFACTOR 106419 97309 0.20 7
DEVELOPMENTAL STAGE 9691 9691 0.02 16
DISEASE 5134 3440 0.01 21
DISRUPTION PHENOTYPE 5045 5045 0.01 22
DOMAIN 38259 33859 0.07 10
ENZYME REGULATION 10841 10841 0.02 15
FUNCTION 416405 399296 0.77 2
INDUCTION 14616 14616 0.03 14
INTERACTION 9601 9601 0.02 17
MASS SPECTROMETRY 5343 4073 0.01 20
MISCELLANEOUS 32311 29870 0.06 12
PATHWAY 131430 119277 0.24 6
PHARMACEUTICAL 85 85 <0.01 29
POLYMORPHISM 876 823 <0.01 24
PTM 44519 35135 0.08 8
RNA EDITING 623 623 <0.01 25
SEQUENCE CAUTION 40704 40704 0.08 9
SIMILARITY 638064 513536 1.19 1
SUBCELLULAR LOCATION 324857 319065 0.60 3
SUBUNIT 240928 240928 0.45 5
TISSUE SPECIFICITY 37920 37920 0.07 11
TOXIC DOSE 510 495 <0.01 27
WEB RESOURCE 8854 7084 0.02 18
Total number of comment topics: 29
Total Number of Average
Line type / subtype number entries per entry Rank
------------------------------------ -------- --------- --------- ----
Features (FT) 3667028 6.81
ACT_SITE 139190 85292 0.26 9
BINDING 272605 73809 0.51 4
CA_BIND 3834 1587 0.01 35
CARBOHYD 106764 27346 0.20 15
CHAIN 545155 532346 1.01 1
COILED 20000 13732 0.04 26
COMPBIAS 53963 28565 0.10 18
CONFLICT 126255 44288 0.23 11
CROSSLNK 6614 3859 0.01 34
DISULFID 107396 29175 0.20 14
DNA_BIND 10286 9398 0.02 31
DOMAIN 159725 94793 0.30 8
HELIX 173302 16986 0.32 7
INIT_MET 15301 15301 0.03 27
INTRAMEM 2139 919 <0.01 37
LIPID 11831 7499 0.02 30
METAL 310967 76643 0.58 3
MOD_RES 194070 63478 0.36 5
MOTIF 35440 22924 0.07 24
MUTAGEN 41260 9610 0.08 22
NON_CONS 2028 743 <0.01 38
NON_STD 354 279 <0.01 39
NON_TER 12186 9309 0.02 29
NP_BIND 116169 72454 0.22 13
PEPTIDE 9908 6692 0.02 32
PROPEP 12583 10817 0.02 28
REGION 126210 65168 0.23 12
REPEAT 94031 13911 0.17 16
SIGNAL 37983 37973 0.07 23
SITE 42620 24644 0.08 20
STRAND 181967 16059 0.34 6
TOPO_DOM 129335 26817 0.24 10
TRANSIT 8193 8090 0.02 33
TRANSMEM 354653 73165 0.66 2
TURN 42316 13753 0.08 21
UNSURE 3087 573 0.01 36
VAR_SEQ 43052 18503 0.08 19
VARIANT 84974 16695 0.16 17
ZN_FING 29282 12882 0.05 25
Total number of feature keys: 39
Total Number of Average
Line type / subtype number entries per entry Rank Category
------------------------------------ -------- --------- --------- ---- -------------------------------------------
Cross-references (DR) 15576158 28.94
2DBase-Ecoli 85 85 <0.01 127 2D gel databases
Aarhus/Ghent-2DPAGE 126 96 <0.01 124 2D gel databases
AGD 932 926 <0.01 102 Organism-specific databases
Allergome 1446 897 <0.01 98 Protein family/group databases
ANU-2DPAGE 27 27 <0.01 134 2D gel databases
ArachnoServer 763 755 <0.01 108 Organism-specific databases
ArrayExpress 59926 59926 0.11 43 Gene expression databases
Bgee 39199 39199 0.07 46 Gene expression databases
BindingDB 295 295 <0.01 120 Other
BioCyc 248606 240153 0.46 22 Enzyme and pathway databases
BRENDA 4294 4285 0.01 89 Enzyme and pathway databases
CAZy 7581 6822 0.01 74 Protein family/group databases
CGD 682 660 <0.01 109 Organism-specific databases
CleanEx 30094 29453 0.06 51 Gene expression databases
COMPLUYEAST-2DPAGE 99 98 <0.01 126 2D gel databases
ConoServer 915 833 <0.01 104 Organism-specific databases
Cornea-2DPAGE 67 67 <0.01 128 2D gel databases
CTD 69075 68422 0.13 41 Organism-specific databases
CYGD 5596 5593 0.01 78 Organism-specific databases
dictyBase 4203 4086 0.01 91 Organism-specific databases
DIP 13810 13714 0.03 68 Protein-protein interaction databases
DisProt 398 395 <0.01 117 3D structure databases
DMDM 16765 16762 0.03 64 Polymorphism databases
DNASU 18658 18587 0.03 59 Protocols and materials databases
DOSAC-COBS-2DPAGE 149 147 <0.01 123 2D gel databases
DrugBank 5319 1628 0.01 80 Other
EchoBASE 4167 4163 0.01 92 Organism-specific databases
ECO2DBASE 352 300 <0.01 119 2D gel databases
EcoGene 4292 4290 0.01 90 Organism-specific databases
eggNOG 430090 430090 0.80 9 Phylogenomic databases
EMBL 929954 527394 1.73 3 Sequence databases
Ensembl 79881 48017 0.15 35 Genome annotation databases
EnsemblBacteria 97993 85019 0.18 31 Genome annotation databases
EnsemblFungi 17419 17127 0.03 63 Genome annotation databases
EnsemblMetazoa 11598 8843 0.02 70 Genome annotation databases
EnsemblPlants 15117 13392 0.03 67 Genome annotation databases
EnsemblProtists 4462 4337 0.01 88 Genome annotation databases
euHCVdb 55 44 <0.01 129 Organism-specific databases
EuPathDB 808 807 <0.01 105 Organism-specific databases
EvolutionaryTrace 16512 16512 0.03 65 Other
FlyBase 5864 5490 0.01 77 Organism-specific databases
Gene3D 330441 253538 0.61 17 Family and domain databases
GeneCards 20037 19729 0.04 56 Organism-specific databases
GeneFarm 3078 3067 0.01 94 Organism-specific databases
GeneID 498069 469564 0.93 6 Genome annotation databases
GeneTree 34526 34511 0.06 47 Phylogenomic databases
Genevestigator 66904 66904 0.12 42 Gene expression databases
GenoList 7069 7057 0.01 75 Organism-specific databases
GenomeReviews 377158 357543 0.70 13 Genome annotation databases
GenomeRNAi 21203 21203 0.04 54 Other
GermOnline 41901 41327 0.08 45 Gene expression databases
GlycoSuiteDB 272 272 <0.01 121 PTM databases
GO 2350326 507710 4.37 1 Ontologies
Gramene 4853 4853 0.01 83 Organism-specific databases
H-InvDB 5596 4774 0.01 79 Organism-specific databases
HAMAP 312264 312058 0.58 18 Family and domain databases
HGNC 19815 19656 0.04 57 Organism-specific databases
HOGENOM 384625 384625 0.71 12 Phylogenomic databases
HOVERGEN 75477 75477 0.14 38 Phylogenomic databases
HPA 17640 13724 0.03 61 Organism-specific databases
HSSP 30466 30466 0.06 50 3D structure databases
InParanoid 69470 69470 0.13 40 Phylogenomic databases
IntAct 33930 33930 0.06 48 Protein-protein interaction databases
InterPro 1769886 514871 3.29 2 Family and domain databases
IPI 95378 67175 0.18 32 Sequence databases
KEGG 473526 447533 0.88 8 Genome annotation databases
KO 371542 371157 0.69 14 Phylogenomic databases
LegioList 765 763 <0.01 107 Organism-specific databases
Leproma 671 668 <0.01 110 Organism-specific databases
MaizeGDB 496 491 <0.01 115 Organism-specific databases
MEROPS 10484 10484 0.02 72 Protein family/group databases
MGI 16477 16432 0.03 66 Organism-specific databases
MIM 17821 13538 0.03 60 Organism-specific databases
MINT 17619 17619 0.03 62 Protein-protein interaction databases
NextBio 69701 69701 0.13 39 Other
neXtProt 20084 20084 0.04 55 Organism-specific databases
OGP 377 377 <0.01 118 2D gel databases
OMA 390637 390637 0.73 11 Phylogenomic databases
Orphanet 4478 2566 0.01 87 Organism-specific databases
OrthoDB 78128 78128 0.15 37 Phylogenomic databases
PANTHER 191286 178861 0.36 24 Family and domain databases
Pathway_Interaction_DB 4568 1666 0.01 86 Enzyme and pathway databases
PATRIC 308720 308695 0.57 20 Genome annotation databases
PDB 89369 18697 0.17 34 3D structure databases
PDBsum 89369 18697 0.17 33 3D structure databases
PeptideAtlas 5163 5163 0.01 81 Proteomic databases
PeroxiBase 768 752 <0.01 106 Protein family/group databases
Pfam 711684 495206 1.32 4 Family and domain databases
PharmGKB 18758 18341 0.03 58 Organism-specific databases
PHCI-2DPAGE 250 250 <0.01 122 2D gel databases
PhosphoSite 33567 33567 0.06 49 PTM databases
PhosSite 627 615 <0.01 111 PTM databases
PhylomeDB 26846 26846 0.05 53 Phylogenomic databases
PIR 118224 108103 0.22 28 Sequence databases
PIRSF 98487 98472 0.18 29 Family and domain databases
PMAP-CutDB 1457 1457 <0.01 97 Other
PMMA-2DPAGE 52 52 <0.01 130 2D gel databases
PomBase 5016 4958 0.01 82 Organism-specific databases
PptaseDB 38 38 <0.01 131 Protein family/group databases
PRIDE 79833 79833 0.15 36 Proteomic databases
PRINTS 137353 120147 0.26 26 Family and domain databases
ProDom 29293 29114 0.05 52 Family and domain databases
ProMEX 513 513 <0.01 113 Proteomic databases
PROSITE 480044 303359 0.89 7 Family and domain databases
ProtClustDB 343244 343244 0.64 15 Phylogenomic databases
ProteinModelPortal 429127 429127 0.80 10 3D structure databases
PseudoCAP 1250 1241 <0.01 100 Organism-specific databases
Rat-heart-2DPAGE 28 28 <0.01 133 2D gel databases
Reactome 12221 7551 0.02 69 Enzyme and pathway databases
REBASE 402 402 <0.01 116 Protein family/group databases
RefSeq 521452 469836 0.97 5 Sequence databases
REPRODUCTION-2DPAGE 1256 1035 <0.01 99 2D gel databases
RGD 7724 7720 0.01 73 Organism-specific databases
SGD 6640 6635 0.01 76 Organism-specific databases
Siena-2DPAGE 103 103 <0.01 125 2D gel databases
SMART 167200 125162 0.31 25 Family and domain databases
SMR 218856 218856 0.41 23 3D structure databases
STRING 309313 309310 0.57 19 Protein-protein interaction databases
SUPFAM 330871 261981 0.61 16 Family and domain databases
SWISS-2DPAGE 1182 1181 <0.01 101 2D gel databases
TAIR 11588 11535 0.02 71 Organism-specific databases
TCDB 3647 3632 0.01 93 Protein family/group databases
TIGR 35 33 <0.01 132 Genome annotation databases
TIGRFAMs 289887 269320 0.54 21 Family and domain databases
TubercuList 1982 1946 <0.01 96 Organism-specific databases
UCD-2DPAGE 509 500 <0.01 114 2D gel databases
UCSC 51926 39791 0.10 44 Genome annotation databases
UniGene 98447 89672 0.18 30 Sequence databases
UniPathway 131304 119154 0.24 27 Enzyme and pathway databases
VectorBase 606 588 <0.01 112 Genome annotation databases
World-2DPAGE 919 908 <0.01 103 2D gel databases
WormBase 4798 3880 0.01 84 Organism-specific databases
Xenbase 4717 4712 0.01 85 Organism-specific databases
ZFIN 2775 2775 0.01 95 Organism-specific databases
Total number of cross-referenced databases: 134
6. AMINO ACID COMPOSITION
6.1 Composition in percent for the complete database
Ala (A) 8.25 Gln (Q) 3.93 Leu (L) 9.66 Ser (S) 6.56
Arg (R) 5.53 Glu (E) 6.75 Lys (K) 5.84 Thr (T) 5.34
Asn (N) 4.06 Gly (G) 7.08 Met (M) 2.42 Trp (W) 1.08
Asp (D) 5.45 His (H) 2.27 Phe (F) 3.86 Tyr (Y) 2.92
Cys (C) 1.37 Ile (I) 5.96 Pro (P) 4.70 Val (V) 6.87
Asx (B) 0.000 Glx (Z) 0.000 Xaa (X) 0.00
The average sequence length in UniProtKB/Swiss-Prot is 355 amino acids.
The shortest sequence is GWA_SEPOF (P83570): 2 amino acids.
The longest sequence is TITIN_MOUSE (A2ASS6): 35213 amino acids.
4. JOURNAL CITATIONS
Note: the following citation statistics reflect the number of distinct
journal citations.
Total number of journals cited in this release of UniProtKB/Swiss-Prot: 2280
4.1 Table of the frequency of journal citations
Journals cited 1x: 751
2x: 290
3x: 152
4x: 118
5x: 84
6x: 69
7x: 57
8x: 45
9x: 32
10x: 29
11- 20x: 182
21- 50x: 190
51-100x: 100
>100x: 181
4.2 List of the most cited journals in UniProtKB/Swiss-Prot
Nb Citations Journal name
-- --------- -------------------------------------------------------------
1 20577 Journal of Biological Chemistry
2 9326 Proceedings of the National Academy of Sciences of the U.S.A.
3 5604 Journal of Bacteriology
4 4973 Biochemical and Biophysical Research Communications
5 4585 Gene
6 4500 Nucleic Acids Research
7 4342 Biochemistry
8 4299 FEBS Letters
9 4155 The EMBO Journal
10 3868 Molecular and Cellular Biology
11 3636 Nature
12 3479 Journal of Molecular Biology
13 3226 European Journal of Biochemistry
14 3180 Biochimica et Biophysica Acta
15 2976 Cell
16 2503 Genomics
17 2463 Journal of Virology
18 2428 Biochemical Journal
19 2399 Science
20 1989 Molecular Microbiology
21 1822 Journal of Cell Biology
22 1769 Plant Physiology
23 1614 Plant Molecular Biology
24 1559 The American Journal of Human Genetics
25 1553 Genes and Development
26 1513 Virology
27 1476 Nature Genetics
28 1433 Human Molecular Genetics
29 1405 Oncogene
30 1331 Molecular and General Genetics
31 1304 Development
32 1290 Human Mutation
33 1255 Molecular Biology of the Cell
34 1225 The Plant Cell
35 1224 Journal of Biochemistry
36 1158 Journal of Immunology
37 1108 Molecular Cell
38 1095 The Plant Journal
39 1090 Genetics
40 1046 Structure
41 1013 Journal of General Virology
42 957 Blood
43 929 Infection and Immunity
44 918 Archives of Biochemistry and Biophysics
45 909 Journal of Cell Science
46 831 Microbiology
47 816 Developmental Biology
48 794 Cancer Research
49 783 Yeast
50 772 Current Biology
51 715 FEMS Microbiology Letters
52 649 Acta Crystallographica, Section D
53 636 Protein Science
54 629 Journal of Neuroscience
55 623 Applied and Environmental Microbiology
56 623 Toxicon
57 622 Human Genetics
58 618 Nature Structural Biology
59 616 Mechanisms of Development
60 579 Neuron
61 565 Journal of Clinical Investigation
62 537 Current Genetics
63 532 American Journal of Physiology
64 522 The Journal of Experimental Medicine
65 483 Proteins
66 481 Molecular Endocrinology
67 479 Mammalian Genome
68 457 Journal of Neurochemistry
69 457 PLoS ONE
70 456 Immunogenetics
71 441 The Journal of Clinical Endocrinology and Metabolism
72 441 Plant and Cell Physiology
73 433 Bioscience, Biotechnology, and Biochemistry
74 430 Endocrinology
75 428 Molecular and Biochemical Parasitology
76 423 Nature Cell Biology
77 400 Journal of Medical Genetics
78 395 Journal of Molecular Evolution
79 377 DNA and Cell Biology
80 375 Molecular Biology and Evolution
81 367 Experimental Cell Research
82 364 DNA Sequence
83 342 Peptides
84 326 Brain Research. Molecular Brain Research
85 324 Tissue Antigens
86 318 Comparative Biochemistry and Physiology
87 311 Developmental Cell
88 310 RNA
89 310 Antimicrobial Agents and Chemotherapy
90 303 Journal of Investigative Dermatology
91 302 Molecular Pharmacology
92 294 Biological Chemistry Hoppe-Seyler
93 293 Nature Structural and Molecular Biology
94 290 Planta
95 288 The FEBS Journal
96 280 Biology of Reproduction
97 279 Cytogenetics and Cell Genetics
98 277 Neurology
99 266 Developmental Dynamics
100 263 Genome Research
101 262 Virus Research
102 254 Journal of General Microbiology
103 248 EMBO Reports
104 245 Immunity
105 243 Molecular Plant-Microbe Interactions
106 243 Biochimie
107 235 Eukaryotic Cell
108 233 The New England Journal of Medicine
109 232 The FASEB Journal
110 232 Genes to Cells
111 227 European Journal of Immunology
112 222 Annals of Neurology
113 219 European Journal of Human Genetics
114 218 Hoppe-Seyler's Zeitschrift fur Physiologische Chemie
115 212 DNA Research
116 209 Journal of Human Genetics
117 197 Investigative Ophthalmology and Visual Science
118 196 Archives of Microbiology
119 195 Journal of the American Chemical Society
120 188 Nature Immunology
121 188 Archives of Virology
122 184 Journal of Cellular Biochemistry
123 183 Molecular and Cellular Endocrinology
124 183 American Journal of Medical Genetics. Part A
125 182 BMC Genomics
126 180 Molecular Immunology
127 177 Glycobiology
128 176 Clinical Genetics
129 172 Insect Biochemistry and Molecular Biology
130 171 Diabetes
131 168 American Journal of Medical Genetics
132 167 Journal of Medicinal Chemistry
133 167 Molecular Phylogenetics and Evolution
134 165 Acta Crystallographica, Section F
135 165 Journal of Experimental Botany
136 163 Circulation Research
137 159 International Journal of Cancer
138 159 DNA
139 158 Molecular Reproduction and Development
140 155 Hemoglobin
141 154 Bioorganicheskaia Khimiia
142 154 Molecular Genetics and Metabolism
143 150 PLoS Genetics
144 149 Phytochemistry
145 149 Molecular and Cellular Neuroscience
146 149 Biological Chemistry
147 148 Molecular Genetics and Genomics
148 145 Journal of Lipid Research
149 145 Protein Expression and Purification
150 144 Traffic
5. STATISTICS FOR SOME LINE TYPES
The following table summarizes the total number of some UniProtKB/Swiss-Prot lines,
as well as the number of entries with at least one such line, and the
frequency of the lines.
Total Number of Average
Line type / subtype number entries per entry
------------------------------------ -------- --------- ---------
References (RL) 1028777 1.91
Journal 822062 413904 1.53 1
Submitted to EMBL/GenBank/DDBJ 197677 177199 0.37 2
Submitted to other databases 6851 6388 0.01 3
Book citation 717 703 <0.01 4
Plant Gene Register 578 566 <0.01 5
Thesis 413 410 <0.01 6
Unpublished observations 283 279 <0.01 7
Patent 190 187 <0.01 8
Worm Breeder's Gazette 6 6 <0.01 9
Total number of distinct authors cited in UniProtKB/Swiss-Prot: 328522
Total Number of Average
Line type / subtype number entries per entry Rank
------------------------------------ -------- --------- --------- ----
Comments (CC) 2400283 4.46
ALLERGEN 520 520 <0.01 26
ALTERNATIVE PRODUCTS 21332 21332 0.04 13
BIOPHYSICOCHEMICAL PROPERTIES 4624 4624 0.01 23
BIOTECHNOLOGY 332 330 <0.01 28
CATALYTIC ACTIVITY 242353 218947 0.45 4
CAUTION 8087 7921 0.02 19
COFACTOR 106419 97309 0.20 7
DEVELOPMENTAL STAGE 9691 9691 0.02 16
DISEASE 5134 3440 0.01 21
DISRUPTION PHENOTYPE 5045 5045 0.01 22
DOMAIN 38259 33859 0.07 10
ENZYME REGULATION 10841 10841 0.02 15
FUNCTION 416405 399296 0.77 2
INDUCTION 14616 14616 0.03 14
INTERACTION 9601 9601 0.02 17
MASS SPECTROMETRY 5343 4073 0.01 20
MISCELLANEOUS 32311 29870 0.06 12
PATHWAY 131430 119277 0.24 6
PHARMACEUTICAL 85 85 <0.01 29
POLYMORPHISM 876 823 <0.01 24
PTM 44519 35135 0.08 8
RNA EDITING 623 623 <0.01 25
SEQUENCE CAUTION 40704 40704 0.08 9
SIMILARITY 638064 513536 1.19 1
SUBCELLULAR LOCATION 324857 319065 0.60 3
SUBUNIT 240928 240928 0.45 5
TISSUE SPECIFICITY 37920 37920 0.07 11
TOXIC DOSE 510 495 <0.01 27
WEB RESOURCE 8854 7084 0.02 18
Total number of comment topics: 29
Total Number of Average
Line type / subtype number entries per entry Rank
------------------------------------ -------- --------- --------- ----
Features (FT) 3667028 6.81
ACT_SITE 139190 85292 0.26 9
BINDING 272605 73809 0.51 4
CA_BIND 3834 1587 0.01 35
CARBOHYD 106764 27346 0.20 15
CHAIN 545155 532346 1.01 1
COILED 20000 13732 0.04 26
COMPBIAS 53963 28565 0.10 18
CONFLICT 126255 44288 0.23 11
CROSSLNK 6614 3859 0.01 34
DISULFID 107396 29175 0.20 14
DNA_BIND 10286 9398 0.02 31
DOMAIN 159725 94793 0.30 8
HELIX 173302 16986 0.32 7
INIT_MET 15301 15301 0.03 27
INTRAMEM 2139 919 <0.01 37
LIPID 11831 7499 0.02 30
METAL 310967 76643 0.58 3
MOD_RES 194070 63478 0.36 5
MOTIF 35440 22924 0.07 24
MUTAGEN 41260 9610 0.08 22
NON_CONS 2028 743 <0.01 38
NON_STD 354 279 <0.01 39
NON_TER 12186 9309 0.02 29
NP_BIND 116169 72454 0.22 13
PEPTIDE 9908 6692 0.02 32
PROPEP 12583 10817 0.02 28
REGION 126210 65168 0.23 12
REPEAT 94031 13911 0.17 16
SIGNAL 37983 37973 0.07 23
SITE 42620 24644 0.08 20
STRAND 181967 16059 0.34 6
TOPO_DOM 129335 26817 0.24 10
TRANSIT 8193 8090 0.02 33
TRANSMEM 354653 73165 0.66 2
TURN 42316 13753 0.08 21
UNSURE 3087 573 0.01 36
VAR_SEQ 43052 18503 0.08 19
VARIANT 84974 16695 0.16 17
ZN_FING 29282 12882 0.05 25
Total number of feature keys: 39
Total Number of Average
Line type / subtype number entries per entry Rank Category
------------------------------------ -------- --------- --------- ---- -------------------------------------------
Cross-references (DR) 15576158 28.94
2DBase-Ecoli 85 85 <0.01 127 2D gel databases
Aarhus/Ghent-2DPAGE 126 96 <0.01 124 2D gel databases
AGD 932 926 <0.01 102 Organism-specific databases
Allergome 1446 897 <0.01 98 Protein family/group databases
ANU-2DPAGE 27 27 <0.01 134 2D gel databases
ArachnoServer 763 755 <0.01 108 Organism-specific databases
ArrayExpress 59926 59926 0.11 43 Gene expression databases
Bgee 39199 39199 0.07 46 Gene expression databases
BindingDB 295 295 <0.01 120 Other
BioCyc 248606 240153 0.46 22 Enzyme and pathway databases
BRENDA 4294 4285 0.01 89 Enzyme and pathway databases
CAZy 7581 6822 0.01 74 Protein family/group databases
CGD 682 660 <0.01 109 Organism-specific databases
CleanEx 30094 29453 0.06 51 Gene expression databases
COMPLUYEAST-2DPAGE 99 98 <0.01 126 2D gel databases
ConoServer 915 833 <0.01 104 Organism-specific databases
Cornea-2DPAGE 67 67 <0.01 128 2D gel databases
CTD 69075 68422 0.13 41 Organism-specific databases
CYGD 5596 5593 0.01 78 Organism-specific databases
dictyBase 4203 4086 0.01 91 Organism-specific databases
DIP 13810 13714 0.03 68 Protein-protein interaction databases
DisProt 398 395 <0.01 117 3D structure databases
DMDM 16765 16762 0.03 64 Polymorphism databases
DNASU 18658 18587 0.03 59 Protocols and materials databases
DOSAC-COBS-2DPAGE 149 147 <0.01 123 2D gel databases
DrugBank 5319 1628 0.01 80 Other
EchoBASE 4167 4163 0.01 92 Organism-specific databases
ECO2DBASE 352 300 <0.01 119 2D gel databases
EcoGene 4292 4290 0.01 90 Organism-specific databases
eggNOG 430090 430090 0.80 9 Phylogenomic databases
EMBL 929954 527394 1.73 3 Sequence databases
Ensembl 79881 48017 0.15 35 Genome annotation databases
EnsemblBacteria 97993 85019 0.18 31 Genome annotation databases
EnsemblFungi 17419 17127 0.03 63 Genome annotation databases
EnsemblMetazoa 11598 8843 0.02 70 Genome annotation databases
EnsemblPlants 15117 13392 0.03 67 Genome annotation databases
EnsemblProtists 4462 4337 0.01 88 Genome annotation databases
euHCVdb 55 44 <0.01 129 Organism-specific databases
EuPathDB 808 807 <0.01 105 Organism-specific databases
EvolutionaryTrace 16512 16512 0.03 65 Other
FlyBase 5864 5490 0.01 77 Organism-specific databases
Gene3D 330441 253538 0.61 17 Family and domain databases
GeneCards 20037 19729 0.04 56 Organism-specific databases
GeneFarm 3078 3067 0.01 94 Organism-specific databases
GeneID 498069 469564 0.93 6 Genome annotation databases
GeneTree 34526 34511 0.06 47 Phylogenomic databases
Genevestigator 66904 66904 0.12 42 Gene expression databases
GenoList 7069 7057 0.01 75 Organism-specific databases
GenomeReviews 377158 357543 0.70 13 Genome annotation databases
GenomeRNAi 21203 21203 0.04 54 Other
GermOnline 41901 41327 0.08 45 Gene expression databases
GlycoSuiteDB 272 272 <0.01 121 PTM databases
GO 2350326 507710 4.37 1 Ontologies
Gramene 4853 4853 0.01 83 Organism-specific databases
H-InvDB 5596 4774 0.01 79 Organism-specific databases
HAMAP 312264 312058 0.58 18 Family and domain databases
HGNC 19815 19656 0.04 57 Organism-specific databases
HOGENOM 384625 384625 0.71 12 Phylogenomic databases
HOVERGEN 75477 75477 0.14 38 Phylogenomic databases
HPA 17640 13724 0.03 61 Organism-specific databases
HSSP 30466 30466 0.06 50 3D structure databases
InParanoid 69470 69470 0.13 40 Phylogenomic databases
IntAct 33930 33930 0.06 48 Protein-protein interaction databases
InterPro 1769886 514871 3.29 2 Family and domain databases
IPI 95378 67175 0.18 32 Sequence databases
KEGG 473526 447533 0.88 8 Genome annotation databases
KO 371542 371157 0.69 14 Phylogenomic databases
LegioList 765 763 <0.01 107 Organism-specific databases
Leproma 671 668 <0.01 110 Organism-specific databases
MaizeGDB 496 491 <0.01 115 Organism-specific databases
MEROPS 10484 10484 0.02 72 Protein family/group databases
MGI 16477 16432 0.03 66 Organism-specific databases
MIM 17821 13538 0.03 60 Organism-specific databases
MINT 17619 17619 0.03 62 Protein-protein interaction databases
NextBio 69701 69701 0.13 39 Other
neXtProt 20084 20084 0.04 55 Organism-specific databases
OGP 377 377 <0.01 118 2D gel databases
OMA 390637 390637 0.73 11 Phylogenomic databases
Orphanet 4478 2566 0.01 87 Organism-specific databases
OrthoDB 78128 78128 0.15 37 Phylogenomic databases
PANTHER 191286 178861 0.36 24 Family and domain databases
Pathway_Interaction_DB 4568 1666 0.01 86 Enzyme and pathway databases
PATRIC 308720 308695 0.57 20 Genome annotation databases
PDB 89369 18697 0.17 34 3D structure databases
PDBsum 89369 18697 0.17 33 3D structure databases
PeptideAtlas 5163 5163 0.01 81 Proteomic databases
PeroxiBase 768 752 <0.01 106 Protein family/group databases
Pfam 711684 495206 1.32 4 Family and domain databases
PharmGKB 18758 18341 0.03 58 Organism-specific databases
PHCI-2DPAGE 250 250 <0.01 122 2D gel databases
PhosphoSite 33567 33567 0.06 49 PTM databases
PhosSite 627 615 <0.01 111 PTM databases
PhylomeDB 26846 26846 0.05 53 Phylogenomic databases
PIR 118224 108103 0.22 28 Sequence databases
PIRSF 98487 98472 0.18 29 Family and domain databases
PMAP-CutDB 1457 1457 <0.01 97 Other
PMMA-2DPAGE 52 52 <0.01 130 2D gel databases
PomBase 5016 4958 0.01 82 Organism-specific databases
PptaseDB 38 38 <0.01 131 Protein family/group databases
PRIDE 79833 79833 0.15 36 Proteomic databases
PRINTS 137353 120147 0.26 26 Family and domain databases
ProDom 29293 29114 0.05 52 Family and domain databases
ProMEX 513 513 <0.01 113 Proteomic databases
PROSITE 480044 303359 0.89 7 Family and domain databases
ProtClustDB 343244 343244 0.64 15 Phylogenomic databases
ProteinModelPortal 429127 429127 0.80 10 3D structure databases
PseudoCAP 1250 1241 <0.01 100 Organism-specific databases
Rat-heart-2DPAGE 28 28 <0.01 133 2D gel databases
Reactome 12221 7551 0.02 69 Enzyme and pathway databases
REBASE 402 402 <0.01 116 Protein family/group databases
RefSeq 521452 469836 0.97 5 Sequence databases
REPRODUCTION-2DPAGE 1256 1035 <0.01 99 2D gel databases
RGD 7724 7720 0.01 73 Organism-specific databases
SGD 6640 6635 0.01 76 Organism-specific databases
Siena-2DPAGE 103 103 <0.01 125 2D gel databases
SMART 167200 125162 0.31 25 Family and domain databases
SMR 218856 218856 0.41 23 3D structure databases
STRING 309313 309310 0.57 19 Protein-protein interaction databases
SUPFAM 330871 261981 0.61 16 Family and domain databases
SWISS-2DPAGE 1182 1181 <0.01 101 2D gel databases
TAIR 11588 11535 0.02 71 Organism-specific databases
TCDB 3647 3632 0.01 93 Protein family/group databases
TIGR 35 33 <0.01 132 Genome annotation databases
TIGRFAMs 289887 269320 0.54 21 Family and domain databases
TubercuList 1982 1946 <0.01 96 Organism-specific databases
UCD-2DPAGE 509 500 <0.01 114 2D gel databases
UCSC 51926 39791 0.10 44 Genome annotation databases
UniGene 98447 89672 0.18 30 Sequence databases
UniPathway 131304 119154 0.24 27 Enzyme and pathway databases
VectorBase 606 588 <0.01 112 Genome annotation databases
World-2DPAGE 919 908 <0.01 103 2D gel databases
WormBase 4798 3880 0.01 84 Organism-specific databases
Xenbase 4717 4712 0.01 85 Organism-specific databases
ZFIN 2775 2775 0.01 95 Organism-specific databases
Total number of cross-referenced databases: 134
6. AMINO ACID COMPOSITION
6.1 Composition in percent for the complete database
Ala (A) 8.25 Gln (Q) 3.93 Leu (L) 9.66 Ser (S) 6.56
Arg (R) 5.53 Glu (E) 6.75 Lys (K) 5.84 Thr (T) 5.34
Asn (N) 4.06 Gly (G) 7.08 Met (M) 2.42 Trp (W) 1.08
Asp (D) 5.45 His (H) 2.27 Phe (F) 3.86 Tyr (Y) 2.92
Cys (C) 1.37 Ile (I) 5.96 Pro (P) 4.70 Val (V) 6.87
Asx (B) 0.000 Glx (Z) 0.000 Xaa (X) 0.00
 Legend: gray = aliphatic, red = acidic, green = small hydroxy,
blue = basic, black = aromatic, white = amide, yellow = sulfur
6.2 Classification of the amino acids by their frequency
Leu, Ala, Gly, Val, Glu, Ser, Ile, Lys, Arg, Asp, Thr, Pro, Asn, Gln,
Phe, Tyr, Met, His, Cys, Trp
7. MISCELLANEOUS STATISTICS
4460 entries are encoded on a mitochondrion, and 3687 are encoded on a plasmid.
12188 entries are encoded on a plastid,
of which 21 are encoded on apicoplasts,
11623 on chloroplasts,
51 on organellar chromatophores,
145 on cyanelles,
149 on non-photosynthetic plastids and
199 on unspecified types of plastid.
Number of entries with at least one sequence correction: 74058
Legend: gray = aliphatic, red = acidic, green = small hydroxy,
blue = basic, black = aromatic, white = amide, yellow = sulfur
6.2 Classification of the amino acids by their frequency
Leu, Ala, Gly, Val, Glu, Ser, Ile, Lys, Arg, Asp, Thr, Pro, Asn, Gln,
Phe, Tyr, Met, His, Cys, Trp
7. MISCELLANEOUS STATISTICS
4460 entries are encoded on a mitochondrion, and 3687 are encoded on a plasmid.
12188 entries are encoded on a plastid,
of which 21 are encoded on apicoplasts,
11623 on chloroplasts,
51 on organellar chromatophores,
145 on cyanelles,
149 on non-photosynthetic plastids and
199 on unspecified types of plastid.
Number of entries with at least one sequence correction: 74058