UniProtKB/TrEMBL PROTEIN DATABASE RELEASE 2013_01 STATISTICS
1. INTRODUCTION
Release 2013_01 of 09-Jan-2013 of UniProtKB/TrEMBL contains 29266939 sequence entries,
comprising 9427157298 amino acids .
936120 sequences have been added since release 2012_11, the sequence data of
36647 existing entries has been updated and the annotations of
11841242 entries have been revised. This represents an increase of 3%.
Number of fragments: 3804361
Protein existence (PE): entries %
1: Evidence at protein level 14128 0.05%
2: Evidence at transcript level 647041 2.21%
3: Inferred from homology 6777086 23.16%
4: Predicted 21828684 74.58%
5: Uncertain 0 0.00%
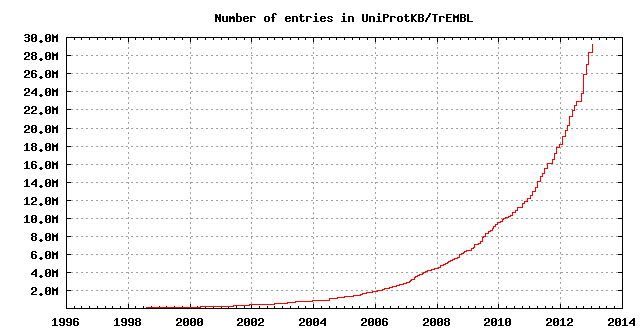
The growth of the database is summarized below.
 2. TAXONOMIC ORIGIN
Total number of species represented in this release of UniProtKB/TrEMBL: 386951
The first twenty species represent 1753638 sequences: 6 % of the
total number of entries.
2.1 Table of the frequency of occurrence of species
Species represented 1x:16242
2x:64555
3x:34983
4x:23250
5x:14696
6x:10625
7x: 7987
8x: 6241
9x: 5054
10x: 9820
11- 20x:25669
21- 50x: 9048
51-100x: 3468
>100x: 9130
2.2 Table of the most represented species
------ --------- --------------------------------------------
Number Frequency Species
------ --------- --------------------------------------------
1 502941 Human immunodeficiency virus 1
2 180820 uncultured bacterium
3 113576 Homo sapiens (Human)
4 96951 Oryza sativa subsp. japonica (Rice)
5 78428 Hepatitis C virus
6 73724 Glycine max (Soybean) (Glycine hispida)
7 68970 Macaca mulatta (Rhesus macaque)
8 58232 Mus musculus (Mouse)
9 56116 Medicago truncatula (Barrel medic) (Medicago tribuloides)
10 54935 Hepatitis B virus (HBV)
11 54191 Danio rerio (Zebrafish) (Brachydanio rerio)
12 54089 Vitis vinifera (Grape)
13 50594 Trichomonas vaginalis
14 49230 Tetraodon nigroviridis (Spotted green pufferfish) (Chelonodon nigroviridis)
15 48878 Takifugu rubripes (Japanese pufferfish) (Fugu rubripes)
16 44540 Populus trichocarpa (Western balsam poplar)
17 43145 Callithrix jacchus (White-tufted-ear marmoset)
18 42299 Arabidopsis thaliana (Mouse-ear cress)
19 42129 Zea mays (Maize)
20 39850 Paramecium tetraurelia
21 39805 Oryza sativa subsp. indica (Rice)
22 39291 Setaria italica (Foxtail millet) (Panicum italicum)
23 38163 human gut metagenome
24 35879 Solanum lycopersicum (Tomato) (Lycopersicon esculentum)
25 35602 Ailuropoda melanoleuca (Giant panda)
26 35193 Acyrthosiphon pisum (Pea aphid)
27 35066 Caenorhabditis japonica
28 34802 Physcomitrella patens subsp. patens (Moss)
29 34453 Thalassiosira oceanica (Marine diatom)
30 34176 Drosophila melanogaster (Fruit fly)
31 33910 Rattus norvegicus (Rat)
32 33777 Sorghum bicolor (Sorghum) (Sorghum vulgare)
33 33267 Selaginella moellendorffii (Spikemoss)
34 32769 Arabidopsis lyrata subsp. lyrata (Lyre-leaved rock-cress)
35 32339 Oryza brachyantha
36 32122 Caenorhabditis remanei (Caenorhabditis vulgaris)
37 32093 Oryza glaberrima (African rice)
38 31833 Pan troglodytes (Chimpanzee)
39 31472 Sus scrofa (Pig)
40 31397 Ricinus communis (Castor bean)
41 30917 Daphnia pulex (Water flea)
42 30300 Caenorhabditis brenneri (Nematode worm)
43 30143 Brachypodium distachyon (Purple false brome) (Trachynia distachya)
44 29815 Amphimedon queenslandica (Sponge)
45 29451 Strongylocentrotus purpuratus (Purple sea urchin)
46 29315 Pristionchus pacificus
47 29178 Branchiostoma floridae (Florida lancelet) (Amphioxus)
48 29053 Oikopleura dioica (Tunicate)
49 28521 Xenopus tropicalis (Western clawed frog) (Silurana tropicalis)
50 28446 Canis familiaris (Dog) (Canis lupus familiaris)
51 28362 Escherichia coli
52 28343 Simian immunodeficiency virus (SIV)
53 28055 Gasterosteus aculeatus (Three-spined stickleback)
54 27685 Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
55 27490 Oreochromis niloticus (Nile tilapia) (Tilapia nilotica)
56 27089 Gorilla gorilla gorilla (Lowland gorilla)
57 26818 Crassostrea gigas (Pacific oyster) (Crassostrea angulata)
58 26790 Gallus gallus (Chicken)
59 25900 Oryzias latipes (Medaka fish) (Japanese ricefish)
60 25758 Loxodonta africana (African elephant)
61 25721 Phytophthora sojae (strain P6497) (Soybean stem and root rot agent)
62 25384 Bos taurus (Bovine)
63 25074 Oryctolagus cuniculus (Rabbit)
64 24879 Nematostella vectensis (Starlet sea anemone)
65 24643 Tetrahymena thermophila (strain SB210)
66 24200 Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus)
67 24059 Equus caballus (Horse)
68 23715 Ornithorhynchus anatinus (Duckbill platypus)
69 23565 Oxytricha trifallax
70 23225 Nomascus leucogenys (Northern white-cheeked gibbon) (Hylobates leucogenys)
71 23115 Perkinsus marinus (strain ATCC 50983 / TXsc)
72 22714 Monodelphis domestica (Gray short-tailed opossum)
73 22560 Sarcophilus harrisii (Tasmanian devil) (Sarcophilus laniarius)
74 22437 Caenorhabditis elegans
75 22305 Pongo abelii (Sumatran orangutan) (Pongo pygmaeus abelii)
76 22163 gut metagenome
77 21821 Latimeria chalumnae (West Indian ocean coelacanth)
78 21699 Hordeum vulgare var. distichum (Two-rowed barley)
79 21546 Heterocephalus glaber (Naked mole rat)
80 21339 Caenorhabditis briggsae
81 21086 Ixodes scapularis (Black-legged tick) (Deer tick)
82 20854 Myotis lucifugus (Little brown bat)
83 20732 Pelodiscus sinensis (Chinese softshell turtle) (Trionyx sinensis)
84 20130 Otolemur garnettii (Small-eared galago) (Garnett's greater bushbaby)
85 20114 Ciona savignyi (Pacific transparent sea squirt)
86 20069 Cavia porcellus (Guinea pig)
87 19969 Spermophilus tridecemlineatus (Thirteen-lined ground squirrel)
88 19671 Taeniopygia guttata (Zebra finch) (Poephila guttata)
89 19438 Wuchereria bancrofti
90 19320 Toxoplasma gondii
91 19255 Anolis carolinensis (Green anole) (American chameleon)
92 19200 Trypanosoma cruzi (strain CL Brener)
93 18919 Culex quinquefasciatus (Southern house mosquito) (Culex pungens)
94 18771 mine drainage metagenome
95 18706 Drosophila simulans (Fruit fly)
96 18585 Ciona intestinalis (Transparent sea squirt) (Ascidia intestinalis)
97 18121 Atta cephalotes (Leafcutter ant)
98 17839 Laccaria bicolor (strain S238N-H82 / ATCC MYA-4686) (Bicoloured deceiver)
99 17784 Fusarium oxysporum (strain Fo5176) (Panama disease fungus)
100 17599 Phytophthora infestans (strain T30-4) (Potato late blight fungus)
101 17384 Aedes aegypti (Yellowfever mosquito) (Culex aegypti)
102 17374 Bombyx mori (Silk moth)
103 17277 Nasonia vitripennis (Parasitic wasp)
104 17031 Drosophila yakuba (Fruit fly)
105 17011 Tribolium castaneum (Red flour beetle)
106 16946 Rhizopus delemar (strain RA 99-880 / ATCC MYA-4621 / FGSC 9543 / NRRL 43880)
107 16871 Meleagris gallopavo (Common turkey)
108 16714 Drosophila persimilis (Fruit fly)
109 16643 Fusarium oxysporum f. sp. lycopersici
110 16475 Drosophila pseudoobscura pseudoobscura (Fruit fly)
111 16426 Ectocarpus siliculosus (Brown alga)
112 16345 Botryotinia fuckeliana (strain T4) (Noble rot fungus) (Botrytis cinerea)
113 16306 Danaus plexippus (Monarch butterfly)
114 16263 Trichinella spiralis (Trichina worm)
115 16237 Melampsora larici-populina (strain 98AG31 / pathotype 3-4-7)
116 16188 Drosophila sechellia (Fruit fly)
117 16140 Schistosoma japonicum (Blood fluke)
118 16110 Colletotrichum higginsianum (strain IMI 349063) (Crucifer anthracnose fungus)
119 15930 Hepatitis C virus subtype 1b
120 15816 Plasmodium falciparum
121 15793 Puccinia graminis f. sp. tritici (strain CRL 75-36-700-3 / race SCCL)
122 15762 Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173)
123 15715 Naegleria gruberi (Amoeba)
124 15653 Nectria haematococca (strain 77-13-4 / ATCC MYA-4622 / FGSC 9596 / MPVI)
125 15630 Anopheles gambiae (African malaria mosquito)
126 15563 Phytophthora ramorum (Sudden oak death agent)
127 15420 Drosophila willistoni (Fruit fly)
128 15354 Loa loa (Eye worm) (Filaria loa)
129 15225 Pythium ultimum
130 15173 Hepatitis C virus subtype 1a
131 15143 Drosophila ananassae (Fruit fly)
132 15036 Harpegnathos saltator (Jerdon's jumping ant)
133 14927 Drosophila erecta (Fruit fly)
134 14851 Chlamydomonas reinhardtii (Chlamydomonas smithii)
135 14797 Camponotus floridanus (Florida carpenter ant)
136 14788 Drosophila mojavensis (Fruit fly)
137 14701 Drosophila virilis (Fruit fly)
138 14697 Plasmodium chabaudi
139 14650 Drosophila grimshawi (Fruit fly) (Idiomyia grimshawi)
140 14610 Gaeumannomyces graminis var. tritici (strain R3-111a-1)
141 14417 Volvox carteri (Green alga)
142 14339 Serpula lacrymans var. lacrymans (strain S7.3) (Dry rot fungus)
143 14336 Solenopsis invicta (Red imported fire ant) (Solenopsis wagneri)
144 14236 Sclerotinia sclerotiorum (strain ATCC 18683 / 1980 / Ss-1) (White mold)
145 13966 Acromyrmex echinatior (Panamanian leafcutter ant)
146 13867 Phanerochaete carnosa (strain HHB-10118-sp) (White-rot fungus)
147 13863 Clonorchis sinensis (Chinese liver fluke)
148 13801 Macrophomina phaseolina (strain MS6) (Charcoal rot fungus)
149 13766 Aspergillus niger (strain CBS 513.88 / FGSC A1513)
150 13648 Moniliophthora perniciosa (strain FA553 / isolate CP02)
151 13530 Trypanosoma cruzi
152 13329 Aspergillus flavus
153 13266 Coprinopsis cinerea (strain Okayama-7 / 130 / ATCC MYA-4618 / FGSC 9003)
154 13186 Mustela putorius furo (European domestic ferret) (Mustela furo)
155 13121 Schizophyllum commune (strain H4-8 / FGSC 9210) (Split gill fungus)
156 13043 Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958)
157 12983 Albugo laibachii Nc14
158 12950 Stigmatella aurantiaca (strain DW4/3-1)
159 12936 Talaromyces stipitatus (strain ATCC 10500 / CBS 375.48 / QM 6759 / NRRL 1006)
160 12900 Gibberella zeae (strain PH-1 / ATCC MYA-4620 / FGSC 9075 / NRRL 31084)
161 12722 Serpula lacrymans var. lacrymans (strain S7.9) (Dry rot fungus)
162 12696 Trypanosoma congolense (strain IL3000)
163 12682 Penicillium chrysogenum (strain ATCC 28089 / DSM 1075 / Wisconsin 54-1255)
164 12674 Schistosoma mansoni (Blood fluke)
165 12603 Xenopus laevis (African clawed frog)
166 12570 Ralstonia solanacearum (Pseudomonas solanacearum)
167 12447 Fusarium pseudograminearum (strain CS3096) (Wheat and barley crown-rot fungus)
168 12446 Leptosphaeria maculans (strain JN3 / isolate v23.1.3 / race Av1-4-5-6-7-8)
169 12440 Polysphondylium pallidum (Cellular slime mold)
170 12390 Rabies virus
171 12389 Hypocrea virens (strain Gv29-8 / FGSC 10586) (Gliocladium virens)
172 12352 Dictyostelium purpureum (Slime mold)
173 12329 uncultured archaeon
174 12152 Dictyostelium fasciculatum (strain SH3) (Slime mold)
175 11994 Pyrenophora tritici-repentis (strain Pt-1C-BFP) (Wheat tan spot fungus)
176 11993 Colletotrichum graminicola (strain M1.001 / M2 / FGSC 10212)
177 11946 Porcine reproductive and respiratory syndrome virus (PRRSV)
178 11945 Emericella nidulans
179 11927 Helicobacter pylori (Campylobacter pylori)
180 11914 Apis mellifera (Honeybee)
181 11815 Hypocrea atroviridis (strain ATCC 20476 / IMI 206040) (Trichoderma atroviride)
182 11780 Piriformospora indica (strain DSM 11827)
183 11752 Chondrocladia sp. SMF
2.3 Taxonomic distribution of the sequences
2. TAXONOMIC ORIGIN
Total number of species represented in this release of UniProtKB/TrEMBL: 386951
The first twenty species represent 1753638 sequences: 6 % of the
total number of entries.
2.1 Table of the frequency of occurrence of species
Species represented 1x:16242
2x:64555
3x:34983
4x:23250
5x:14696
6x:10625
7x: 7987
8x: 6241
9x: 5054
10x: 9820
11- 20x:25669
21- 50x: 9048
51-100x: 3468
>100x: 9130
2.2 Table of the most represented species
------ --------- --------------------------------------------
Number Frequency Species
------ --------- --------------------------------------------
1 502941 Human immunodeficiency virus 1
2 180820 uncultured bacterium
3 113576 Homo sapiens (Human)
4 96951 Oryza sativa subsp. japonica (Rice)
5 78428 Hepatitis C virus
6 73724 Glycine max (Soybean) (Glycine hispida)
7 68970 Macaca mulatta (Rhesus macaque)
8 58232 Mus musculus (Mouse)
9 56116 Medicago truncatula (Barrel medic) (Medicago tribuloides)
10 54935 Hepatitis B virus (HBV)
11 54191 Danio rerio (Zebrafish) (Brachydanio rerio)
12 54089 Vitis vinifera (Grape)
13 50594 Trichomonas vaginalis
14 49230 Tetraodon nigroviridis (Spotted green pufferfish) (Chelonodon nigroviridis)
15 48878 Takifugu rubripes (Japanese pufferfish) (Fugu rubripes)
16 44540 Populus trichocarpa (Western balsam poplar)
17 43145 Callithrix jacchus (White-tufted-ear marmoset)
18 42299 Arabidopsis thaliana (Mouse-ear cress)
19 42129 Zea mays (Maize)
20 39850 Paramecium tetraurelia
21 39805 Oryza sativa subsp. indica (Rice)
22 39291 Setaria italica (Foxtail millet) (Panicum italicum)
23 38163 human gut metagenome
24 35879 Solanum lycopersicum (Tomato) (Lycopersicon esculentum)
25 35602 Ailuropoda melanoleuca (Giant panda)
26 35193 Acyrthosiphon pisum (Pea aphid)
27 35066 Caenorhabditis japonica
28 34802 Physcomitrella patens subsp. patens (Moss)
29 34453 Thalassiosira oceanica (Marine diatom)
30 34176 Drosophila melanogaster (Fruit fly)
31 33910 Rattus norvegicus (Rat)
32 33777 Sorghum bicolor (Sorghum) (Sorghum vulgare)
33 33267 Selaginella moellendorffii (Spikemoss)
34 32769 Arabidopsis lyrata subsp. lyrata (Lyre-leaved rock-cress)
35 32339 Oryza brachyantha
36 32122 Caenorhabditis remanei (Caenorhabditis vulgaris)
37 32093 Oryza glaberrima (African rice)
38 31833 Pan troglodytes (Chimpanzee)
39 31472 Sus scrofa (Pig)
40 31397 Ricinus communis (Castor bean)
41 30917 Daphnia pulex (Water flea)
42 30300 Caenorhabditis brenneri (Nematode worm)
43 30143 Brachypodium distachyon (Purple false brome) (Trachynia distachya)
44 29815 Amphimedon queenslandica (Sponge)
45 29451 Strongylocentrotus purpuratus (Purple sea urchin)
46 29315 Pristionchus pacificus
47 29178 Branchiostoma floridae (Florida lancelet) (Amphioxus)
48 29053 Oikopleura dioica (Tunicate)
49 28521 Xenopus tropicalis (Western clawed frog) (Silurana tropicalis)
50 28446 Canis familiaris (Dog) (Canis lupus familiaris)
51 28362 Escherichia coli
52 28343 Simian immunodeficiency virus (SIV)
53 28055 Gasterosteus aculeatus (Three-spined stickleback)
54 27685 Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
55 27490 Oreochromis niloticus (Nile tilapia) (Tilapia nilotica)
56 27089 Gorilla gorilla gorilla (Lowland gorilla)
57 26818 Crassostrea gigas (Pacific oyster) (Crassostrea angulata)
58 26790 Gallus gallus (Chicken)
59 25900 Oryzias latipes (Medaka fish) (Japanese ricefish)
60 25758 Loxodonta africana (African elephant)
61 25721 Phytophthora sojae (strain P6497) (Soybean stem and root rot agent)
62 25384 Bos taurus (Bovine)
63 25074 Oryctolagus cuniculus (Rabbit)
64 24879 Nematostella vectensis (Starlet sea anemone)
65 24643 Tetrahymena thermophila (strain SB210)
66 24200 Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus)
67 24059 Equus caballus (Horse)
68 23715 Ornithorhynchus anatinus (Duckbill platypus)
69 23565 Oxytricha trifallax
70 23225 Nomascus leucogenys (Northern white-cheeked gibbon) (Hylobates leucogenys)
71 23115 Perkinsus marinus (strain ATCC 50983 / TXsc)
72 22714 Monodelphis domestica (Gray short-tailed opossum)
73 22560 Sarcophilus harrisii (Tasmanian devil) (Sarcophilus laniarius)
74 22437 Caenorhabditis elegans
75 22305 Pongo abelii (Sumatran orangutan) (Pongo pygmaeus abelii)
76 22163 gut metagenome
77 21821 Latimeria chalumnae (West Indian ocean coelacanth)
78 21699 Hordeum vulgare var. distichum (Two-rowed barley)
79 21546 Heterocephalus glaber (Naked mole rat)
80 21339 Caenorhabditis briggsae
81 21086 Ixodes scapularis (Black-legged tick) (Deer tick)
82 20854 Myotis lucifugus (Little brown bat)
83 20732 Pelodiscus sinensis (Chinese softshell turtle) (Trionyx sinensis)
84 20130 Otolemur garnettii (Small-eared galago) (Garnett's greater bushbaby)
85 20114 Ciona savignyi (Pacific transparent sea squirt)
86 20069 Cavia porcellus (Guinea pig)
87 19969 Spermophilus tridecemlineatus (Thirteen-lined ground squirrel)
88 19671 Taeniopygia guttata (Zebra finch) (Poephila guttata)
89 19438 Wuchereria bancrofti
90 19320 Toxoplasma gondii
91 19255 Anolis carolinensis (Green anole) (American chameleon)
92 19200 Trypanosoma cruzi (strain CL Brener)
93 18919 Culex quinquefasciatus (Southern house mosquito) (Culex pungens)
94 18771 mine drainage metagenome
95 18706 Drosophila simulans (Fruit fly)
96 18585 Ciona intestinalis (Transparent sea squirt) (Ascidia intestinalis)
97 18121 Atta cephalotes (Leafcutter ant)
98 17839 Laccaria bicolor (strain S238N-H82 / ATCC MYA-4686) (Bicoloured deceiver)
99 17784 Fusarium oxysporum (strain Fo5176) (Panama disease fungus)
100 17599 Phytophthora infestans (strain T30-4) (Potato late blight fungus)
101 17384 Aedes aegypti (Yellowfever mosquito) (Culex aegypti)
102 17374 Bombyx mori (Silk moth)
103 17277 Nasonia vitripennis (Parasitic wasp)
104 17031 Drosophila yakuba (Fruit fly)
105 17011 Tribolium castaneum (Red flour beetle)
106 16946 Rhizopus delemar (strain RA 99-880 / ATCC MYA-4621 / FGSC 9543 / NRRL 43880)
107 16871 Meleagris gallopavo (Common turkey)
108 16714 Drosophila persimilis (Fruit fly)
109 16643 Fusarium oxysporum f. sp. lycopersici
110 16475 Drosophila pseudoobscura pseudoobscura (Fruit fly)
111 16426 Ectocarpus siliculosus (Brown alga)
112 16345 Botryotinia fuckeliana (strain T4) (Noble rot fungus) (Botrytis cinerea)
113 16306 Danaus plexippus (Monarch butterfly)
114 16263 Trichinella spiralis (Trichina worm)
115 16237 Melampsora larici-populina (strain 98AG31 / pathotype 3-4-7)
116 16188 Drosophila sechellia (Fruit fly)
117 16140 Schistosoma japonicum (Blood fluke)
118 16110 Colletotrichum higginsianum (strain IMI 349063) (Crucifer anthracnose fungus)
119 15930 Hepatitis C virus subtype 1b
120 15816 Plasmodium falciparum
121 15793 Puccinia graminis f. sp. tritici (strain CRL 75-36-700-3 / race SCCL)
122 15762 Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173)
123 15715 Naegleria gruberi (Amoeba)
124 15653 Nectria haematococca (strain 77-13-4 / ATCC MYA-4622 / FGSC 9596 / MPVI)
125 15630 Anopheles gambiae (African malaria mosquito)
126 15563 Phytophthora ramorum (Sudden oak death agent)
127 15420 Drosophila willistoni (Fruit fly)
128 15354 Loa loa (Eye worm) (Filaria loa)
129 15225 Pythium ultimum
130 15173 Hepatitis C virus subtype 1a
131 15143 Drosophila ananassae (Fruit fly)
132 15036 Harpegnathos saltator (Jerdon's jumping ant)
133 14927 Drosophila erecta (Fruit fly)
134 14851 Chlamydomonas reinhardtii (Chlamydomonas smithii)
135 14797 Camponotus floridanus (Florida carpenter ant)
136 14788 Drosophila mojavensis (Fruit fly)
137 14701 Drosophila virilis (Fruit fly)
138 14697 Plasmodium chabaudi
139 14650 Drosophila grimshawi (Fruit fly) (Idiomyia grimshawi)
140 14610 Gaeumannomyces graminis var. tritici (strain R3-111a-1)
141 14417 Volvox carteri (Green alga)
142 14339 Serpula lacrymans var. lacrymans (strain S7.3) (Dry rot fungus)
143 14336 Solenopsis invicta (Red imported fire ant) (Solenopsis wagneri)
144 14236 Sclerotinia sclerotiorum (strain ATCC 18683 / 1980 / Ss-1) (White mold)
145 13966 Acromyrmex echinatior (Panamanian leafcutter ant)
146 13867 Phanerochaete carnosa (strain HHB-10118-sp) (White-rot fungus)
147 13863 Clonorchis sinensis (Chinese liver fluke)
148 13801 Macrophomina phaseolina (strain MS6) (Charcoal rot fungus)
149 13766 Aspergillus niger (strain CBS 513.88 / FGSC A1513)
150 13648 Moniliophthora perniciosa (strain FA553 / isolate CP02)
151 13530 Trypanosoma cruzi
152 13329 Aspergillus flavus
153 13266 Coprinopsis cinerea (strain Okayama-7 / 130 / ATCC MYA-4618 / FGSC 9003)
154 13186 Mustela putorius furo (European domestic ferret) (Mustela furo)
155 13121 Schizophyllum commune (strain H4-8 / FGSC 9210) (Split gill fungus)
156 13043 Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958)
157 12983 Albugo laibachii Nc14
158 12950 Stigmatella aurantiaca (strain DW4/3-1)
159 12936 Talaromyces stipitatus (strain ATCC 10500 / CBS 375.48 / QM 6759 / NRRL 1006)
160 12900 Gibberella zeae (strain PH-1 / ATCC MYA-4620 / FGSC 9075 / NRRL 31084)
161 12722 Serpula lacrymans var. lacrymans (strain S7.9) (Dry rot fungus)
162 12696 Trypanosoma congolense (strain IL3000)
163 12682 Penicillium chrysogenum (strain ATCC 28089 / DSM 1075 / Wisconsin 54-1255)
164 12674 Schistosoma mansoni (Blood fluke)
165 12603 Xenopus laevis (African clawed frog)
166 12570 Ralstonia solanacearum (Pseudomonas solanacearum)
167 12447 Fusarium pseudograminearum (strain CS3096) (Wheat and barley crown-rot fungus)
168 12446 Leptosphaeria maculans (strain JN3 / isolate v23.1.3 / race Av1-4-5-6-7-8)
169 12440 Polysphondylium pallidum (Cellular slime mold)
170 12390 Rabies virus
171 12389 Hypocrea virens (strain Gv29-8 / FGSC 10586) (Gliocladium virens)
172 12352 Dictyostelium purpureum (Slime mold)
173 12329 uncultured archaeon
174 12152 Dictyostelium fasciculatum (strain SH3) (Slime mold)
175 11994 Pyrenophora tritici-repentis (strain Pt-1C-BFP) (Wheat tan spot fungus)
176 11993 Colletotrichum graminicola (strain M1.001 / M2 / FGSC 10212)
177 11946 Porcine reproductive and respiratory syndrome virus (PRRSV)
178 11945 Emericella nidulans
179 11927 Helicobacter pylori (Campylobacter pylori)
180 11914 Apis mellifera (Honeybee)
181 11815 Hypocrea atroviridis (strain ATCC 20476 / IMI 206040) (Trichoderma atroviride)
182 11780 Piriformospora indica (strain DSM 11827)
183 11752 Chondrocladia sp. SMF
2.3 Taxonomic distribution of the sequences
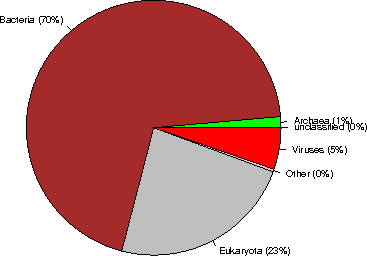 Kingdom sequences (% of the database)
Archaea 408842 ( 1%)
Bacteria 20361597 ( 70%)
Eukaryota 6846383 ( 23%)
Viruses 1547692 ( 5%)
Other 102424 ( <1%)
Within Eukaryota:
Kingdom sequences (% of the database)
Archaea 408842 ( 1%)
Bacteria 20361597 ( 70%)
Eukaryota 6846383 ( 23%)
Viruses 1547692 ( 5%)
Other 102424 ( <1%)
Within Eukaryota:
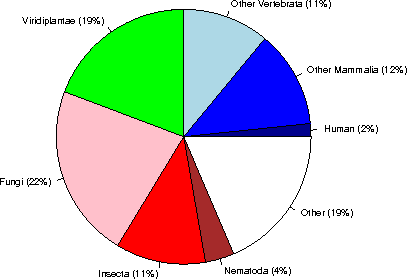 Category sequences (% of Eukaryota) (% of the complete database)
Human 113612 ( 2%) ( 0%)
Other Mammalia 845085 ( 12%) ( 3%)
Other Vertebrata 753401 ( 11%) ( 3%)
Viridiplantae 1317778 ( 19%) ( 5%)
Fungi 1510587 ( 22%) ( 5%)
Insecta 782338 ( 11%) ( 3%)
Nematoda 252562 ( 4%) ( 1%)
Other 1271020 ( 19%) ( 4%)
3. SEQUENCE SIZE
Repartition of the sequences by size (excluding fragments)
From To Number From To Number
1- 50 755730 1001-1100 168936
51- 100 2504899 1101-1200 118418
101- 150 2798075 1201-1300 83249
151- 200 2715002 1301-1400 53034
201- 250 2729580 1401-1500 42871
251- 300 2645019 1501-1600 29671
301- 350 2401964 1601-1700 22537
351- 400 1820598 1701-1800 17087
401- 450 1569876 1801-1900 14194
451- 500 1287032 1901-2000 12118
501- 550 851336 2001-2100 9497
551- 600 655543 2101-2200 9743
601- 650 479546 2201-2300 7592
651- 700 376441 2301-2400 6087
701- 750 317101 2401-2500 5197
751- 800 279539 >2500 42031
801- 850 213365
851- 900 190510
901- 950 132149
951-1000 97011
Category sequences (% of Eukaryota) (% of the complete database)
Human 113612 ( 2%) ( 0%)
Other Mammalia 845085 ( 12%) ( 3%)
Other Vertebrata 753401 ( 11%) ( 3%)
Viridiplantae 1317778 ( 19%) ( 5%)
Fungi 1510587 ( 22%) ( 5%)
Insecta 782338 ( 11%) ( 3%)
Nematoda 252562 ( 4%) ( 1%)
Other 1271020 ( 19%) ( 4%)
3. SEQUENCE SIZE
Repartition of the sequences by size (excluding fragments)
From To Number From To Number
1- 50 755730 1001-1100 168936
51- 100 2504899 1101-1200 118418
101- 150 2798075 1201-1300 83249
151- 200 2715002 1301-1400 53034
201- 250 2729580 1401-1500 42871
251- 300 2645019 1501-1600 29671
301- 350 2401964 1601-1700 22537
351- 400 1820598 1701-1800 17087
401- 450 1569876 1801-1900 14194
451- 500 1287032 1901-2000 12118
501- 550 851336 2001-2100 9497
551- 600 655543 2101-2200 9743
601- 650 479546 2201-2300 7592
651- 700 376441 2301-2400 6087
701- 750 317101 2401-2500 5197
751- 800 279539 >2500 42031
801- 850 213365
851- 900 190510
901- 950 132149
951-1000 97011
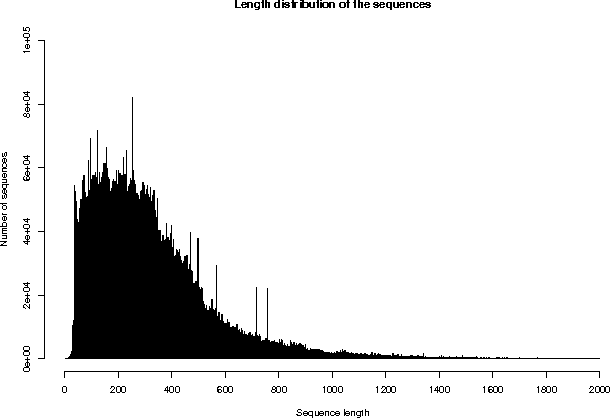 The average sequence length in UniProtKB/TrEMBL is 322 amino acids.
The shortest sequence is G0XMK1_9MYRT: 1 amino acids.
The longest sequence is Q3ASY8_CHLCH: 36805 amino acids.
4. STATISTICS FOR SOME LINE TYPES
The following table summarizes the total number of some UniProtKB/TrEMBL lines,
as well as the number of entries with at least one such line, and the
frequency of the lines.
Total Number of Average
Line type / subtype number entries per entry
--------------------------------- -------- --------- ---------
References (RL) 35659949 1.22
Submitted to EMBL/GenBank/DDBJ 19645186 18124632 0.67
Journal 14507716 13660992 0.50
Submitted to other databases 1490660 1481406 0.05
Thesis 9861 9803 <0.01
Book citation 6506 6457 <0.01
Unpublished observations 19 19 <0.01
Patent 1 1 <0.01
Total number of distinct authors cited in UniProtKB/TrEMBL: 456562
Total Number of Average
Line type / subtype number entries per entry Rank
--------------------------------- -------- --------- --------- ----
Comments (CC) 36180072 1.24
CATALYTIC ACTIVITY 3133293 2851026 0.11 4
CAUTION 13393492 13393247 0.46 1
COFACTOR 1186735 1098197 0.04 8
DOMAIN 118504 113749 <0.01 9
FUNCTION 3425325 3205049 0.12 3
INTERACTION 690 690 <0.01 11
MISCELLANEOUS 82701 82605 <0.01 10
PATHWAY 1552547 1411355 0.05 7
SIMILARITY 8834143 7669541 0.30 2
SUBCELLULAR LOCATION 2759375 2633226 0.09 5
SUBUNIT 1693267 1673876 0.06 6
Total number of comment topics: 11
Total Number of Average
Line type / subtype number entries per entry Rank
--------------------------------- -------- --------- --------- ----
Features (FT) 7381377 0.25
CHAIN 774905 642346 0.03 2
NON_TER 5990131 3805013 0.20 1
SIGNAL 615484 612214 0.02 3
TRANSIT 857 856 <0.01 4
Total number of feature keys: 4
Total Number of Average
Line type / subtype number entries per entry Rank Category
--------------------------------- -------- --------- --------- ---- -------------------------------------------
Cross-references (DR) 330404216 11.29
AGD 2525 2525 <0.01 85 Organism-specific databases
ANU-2DPAGE 52 52 <0.01 101 2D gel databases
Allergome 3024 2409 <0.01 81 Protein family/group databases
ArachnoServer 66 66 <0.01 100 Organism-specific databases
ArrayExpress 86858 86858 <0.01 52 Gene expression databases
BRENDA 2680 2651 <0.01 83 Enzyme and pathway databases
Bgee 118824 118824 <0.01 49 Gene expression databases
BioCyc 3585778 3547066 0.12 20 Enzyme and pathway databases
CAZy 74131 69652 <0.01 56 Protein family/group databases
CGD 7064 7064 <0.01 77 Organism-specific databases
COMPLUYEAST-2DPAGE 5 5 <0.01 107 2D gel databases
CTD 322062 320469 0.01 38 Organism-specific databases
ChEMBL 576 576 <0.01 92 Other
ConoServer 160 160 <0.01 96 Organism-specific databases
DIP 2782 2777 <0.01 82 Protein-protein interaction databases
DNASU 43665 43327 <0.01 61 Protocols and materials databases
EMBL 32113816 28423223 1.10 3 Sequence databases
Ensembl 959177 943419 0.03 29 Genome annotation databases
EnsemblBacteria 834798 800722 0.03 30 Genome annotation databases
EnsemblFungi 262813 261328 0.01 41 Genome annotation databases
EnsemblMetazoa 629747 614533 0.02 32 Genome annotation databases
EnsemblPlants 425229 405703 0.01 37 Genome annotation databases
EnsemblProtists 126697 125197 <0.01 48 Genome annotation databases
EuPathDB 178957 178954 0.01 45 Organism-specific databases
EvolutionaryTrace 8177 8177 <0.01 75 Other
FlyBase 182089 180690 0.01 44 Organism-specific databases
GO 59000915 18239516 2.02 2 Ontologies
Gene3D 12548459 9987923 0.43 6 Family and domain databases
GeneID 8760277 8545494 0.30 9 Genome annotation databases
GeneTree 798347 798164 0.03 31 Phylogenomic databases
Genevestigator 93341 93333 <0.01 51 Gene expression databases
GenoList 14735 14462 <0.01 73 Organism-specific databases
GenomeRNAi 21434 21434 <0.01 67 Other
GenomeReviews 4251570 4152775 0.15 16 Genome annotation databases
Gramene 67608 67608 <0.01 57 Organism-specific databases
H-InvDB 626 478 <0.01 91 Organism-specific databases
HAMAP 2916833 2880798 0.10 22 Family and domain databases
HGNC 48958 48878 <0.01 59 Organism-specific databases
HOGENOM 3658839 3658792 0.13 19 Phylogenomic databases
HOVERGEN 311258 311246 0.01 39 Phylogenomic databases
HSSP 250661 250434 0.01 42 3D structure databases
IPI 309350 308676 0.01 40 Sequence databases
InParanoid 189538 189538 0.01 43 Phylogenomic databases
IntAct 16845 16845 <0.01 71 Protein-protein interaction databases
InterPro 63181437 22759474 2.16 1 Family and domain databases
KEGG 7902138 7711636 0.27 11 Genome annotation databases
KO 3098495 3085017 0.11 21 Phylogenomic databases
LegioList 5138 5110 <0.01 78 Organism-specific databases
Leproma 1272 1270 <0.01 88 Organism-specific databases
MEROPS 81141 81140 <0.01 53 Protein family/group databases
MGI 35077 34422 <0.01 63 Organism-specific databases
MINT 8590 8590 <0.01 74 Protein-protein interaction databases
NextBio 104044 103709 <0.01 50 Other
OMA 3889766 3889385 0.13 18 Phylogenomic databases
OrthoDB 557052 557014 0.02 34 Phylogenomic databases
PANTHER 4184001 3955873 0.14 17 Family and domain databases
PATRIC 8310247 8310154 0.28 10 Genome annotation databases
PDB 18385 10298 <0.01 68 3D structure databases
PDBsum 18152 10143 <0.01 69 3D structure databases
PHCI-2DPAGE 99 99 <0.01 98 2D gel databases
PIR 173697 140856 0.01 46 Sequence databases
PIRSF 2516170 2515493 0.09 26 Family and domain databases
PMAP-CutDB 214 214 <0.01 94 Other
PMMA-2DPAGE 2 2 <0.01 108 2D gel databases
PRIDE 476086 476086 0.02 36 Proteomic databases
PRINTS 4469756 3973785 0.15 15 Family and domain databases
PROSITE 14613027 9689583 0.50 5 Family and domain databases
Pathway_Interaction_DB 11 9 <0.01 106 Enzyme and pathway databases
PaxDb 17042 17042 <0.01 70 Proteomic databases
PeptideAtlas 144 144 <0.01 97 Proteomic databases
PeroxiBase 2558 2550 <0.01 84 Protein family/group databases
Pfam 28789057 21143842 0.98 4 Family and domain databases
PharmGKB 4279 4279 <0.01 80 Organism-specific databases
PhosphoSite 1167 1167 <0.01 89 PTM databases
PhylomeDB 151147 151147 0.01 47 Phylogenomic databases
PomBase 40 27 <0.01 102 Organism-specific databases
PptaseDB 36 34 <0.01 103 Protein family/group databases
ProDom 572760 547986 0.02 33 Family and domain databases
ProMEX 268 268 <0.01 93 Proteomic databases
ProtClustDB 2720788 2720777 0.09 24 Phylogenomic databases
ProteinModelPortal 7763747 7763747 0.27 12 3D structure databases
PseudoCAP 4539 4533 <0.01 79 Organism-specific databases
REBASE 32949 32946 <0.01 64 Protein family/group databases
REPRODUCTION-2DPAGE 84 83 <0.01 99 2D gel databases
RGD 24752 24429 <0.01 66 Organism-specific databases
Reactome 209 179 <0.01 95 Enzyme and pathway databases
RefSeq 8800149 8554497 0.30 8 Sequence databases
SGD 11 11 <0.01 105 Organism-specific databases
SMART 6511821 4935454 0.22 14 Family and domain databases
SMR 1667955 1667955 0.06 27 3D structure databases
STRING 2588334 2588334 0.09 25 Protein-protein interaction databases
SUPFAM 12032612 9897317 0.41 7 Family and domain databases
SWISS-2DPAGE 28 28 <0.01 104 2D gel databases
Siena-2DPAGE 2 2 <0.01 109 2D gel databases
TAIR 15743 15666 <0.01 72 Organism-specific databases
TCDB 2389 2377 <0.01 86 Protein family/group databases
TIGRFAMs 6651588 6065915 0.23 13 Family and domain databases
TubercuList 1991 1986 <0.01 87 Organism-specific databases
UCSC 64222 64050 <0.01 58 Genome annotation databases
UniGene 545475 514039 0.02 35 Sequence databases
UniPathway 1514470 1409900 0.05 28 Enzyme and pathway databases
VectorBase 78249 77732 <0.01 54 Genome annotation databases
World-2DPAGE 675 670 <0.01 90 2D gel databases
WormBase 42337 42219 <0.01 62 Organism-specific databases
Xenbase 25586 25469 <0.01 65 Organism-specific databases
ZFIN 45574 45448 <0.01 60 Organism-specific databases
dictyBase 7996 7774 <0.01 76 Organism-specific databases
eggNOG 2770833 2770812 0.09 23 Phylogenomic databases
euHCVdb 75267 75264 <0.01 55 Organism-specific databases
Number of explicitly cross-referenced databases: 135
5. AMINO ACID COMPOSITION
5.1 Composition in percent for the complete database
Ala (A) 8.63 Gln (Q) 3.97 Leu (L) 9.92 Ser (S) 6.65
Arg (R) 5.42 Glu (E) 6.18 Lys (K) 5.30 Thr (T) 5.56
Asn (N) 4.11 Gly (G) 7.08 Met (M) 2.47 Trp (W) 1.30
Asp (D) 5.32 His (H) 2.21 Phe (F) 4.03 Tyr (Y) 3.05
Cys (C) 1.24 Ile (I) 6.00 Pro (P) 4.67 Val (V) 6.77
Asx (B) 0.000 Glx (Z) 0 Xaa (X) 0.03
The average sequence length in UniProtKB/TrEMBL is 322 amino acids.
The shortest sequence is G0XMK1_9MYRT: 1 amino acids.
The longest sequence is Q3ASY8_CHLCH: 36805 amino acids.
4. STATISTICS FOR SOME LINE TYPES
The following table summarizes the total number of some UniProtKB/TrEMBL lines,
as well as the number of entries with at least one such line, and the
frequency of the lines.
Total Number of Average
Line type / subtype number entries per entry
--------------------------------- -------- --------- ---------
References (RL) 35659949 1.22
Submitted to EMBL/GenBank/DDBJ 19645186 18124632 0.67
Journal 14507716 13660992 0.50
Submitted to other databases 1490660 1481406 0.05
Thesis 9861 9803 <0.01
Book citation 6506 6457 <0.01
Unpublished observations 19 19 <0.01
Patent 1 1 <0.01
Total number of distinct authors cited in UniProtKB/TrEMBL: 456562
Total Number of Average
Line type / subtype number entries per entry Rank
--------------------------------- -------- --------- --------- ----
Comments (CC) 36180072 1.24
CATALYTIC ACTIVITY 3133293 2851026 0.11 4
CAUTION 13393492 13393247 0.46 1
COFACTOR 1186735 1098197 0.04 8
DOMAIN 118504 113749 <0.01 9
FUNCTION 3425325 3205049 0.12 3
INTERACTION 690 690 <0.01 11
MISCELLANEOUS 82701 82605 <0.01 10
PATHWAY 1552547 1411355 0.05 7
SIMILARITY 8834143 7669541 0.30 2
SUBCELLULAR LOCATION 2759375 2633226 0.09 5
SUBUNIT 1693267 1673876 0.06 6
Total number of comment topics: 11
Total Number of Average
Line type / subtype number entries per entry Rank
--------------------------------- -------- --------- --------- ----
Features (FT) 7381377 0.25
CHAIN 774905 642346 0.03 2
NON_TER 5990131 3805013 0.20 1
SIGNAL 615484 612214 0.02 3
TRANSIT 857 856 <0.01 4
Total number of feature keys: 4
Total Number of Average
Line type / subtype number entries per entry Rank Category
--------------------------------- -------- --------- --------- ---- -------------------------------------------
Cross-references (DR) 330404216 11.29
AGD 2525 2525 <0.01 85 Organism-specific databases
ANU-2DPAGE 52 52 <0.01 101 2D gel databases
Allergome 3024 2409 <0.01 81 Protein family/group databases
ArachnoServer 66 66 <0.01 100 Organism-specific databases
ArrayExpress 86858 86858 <0.01 52 Gene expression databases
BRENDA 2680 2651 <0.01 83 Enzyme and pathway databases
Bgee 118824 118824 <0.01 49 Gene expression databases
BioCyc 3585778 3547066 0.12 20 Enzyme and pathway databases
CAZy 74131 69652 <0.01 56 Protein family/group databases
CGD 7064 7064 <0.01 77 Organism-specific databases
COMPLUYEAST-2DPAGE 5 5 <0.01 107 2D gel databases
CTD 322062 320469 0.01 38 Organism-specific databases
ChEMBL 576 576 <0.01 92 Other
ConoServer 160 160 <0.01 96 Organism-specific databases
DIP 2782 2777 <0.01 82 Protein-protein interaction databases
DNASU 43665 43327 <0.01 61 Protocols and materials databases
EMBL 32113816 28423223 1.10 3 Sequence databases
Ensembl 959177 943419 0.03 29 Genome annotation databases
EnsemblBacteria 834798 800722 0.03 30 Genome annotation databases
EnsemblFungi 262813 261328 0.01 41 Genome annotation databases
EnsemblMetazoa 629747 614533 0.02 32 Genome annotation databases
EnsemblPlants 425229 405703 0.01 37 Genome annotation databases
EnsemblProtists 126697 125197 <0.01 48 Genome annotation databases
EuPathDB 178957 178954 0.01 45 Organism-specific databases
EvolutionaryTrace 8177 8177 <0.01 75 Other
FlyBase 182089 180690 0.01 44 Organism-specific databases
GO 59000915 18239516 2.02 2 Ontologies
Gene3D 12548459 9987923 0.43 6 Family and domain databases
GeneID 8760277 8545494 0.30 9 Genome annotation databases
GeneTree 798347 798164 0.03 31 Phylogenomic databases
Genevestigator 93341 93333 <0.01 51 Gene expression databases
GenoList 14735 14462 <0.01 73 Organism-specific databases
GenomeRNAi 21434 21434 <0.01 67 Other
GenomeReviews 4251570 4152775 0.15 16 Genome annotation databases
Gramene 67608 67608 <0.01 57 Organism-specific databases
H-InvDB 626 478 <0.01 91 Organism-specific databases
HAMAP 2916833 2880798 0.10 22 Family and domain databases
HGNC 48958 48878 <0.01 59 Organism-specific databases
HOGENOM 3658839 3658792 0.13 19 Phylogenomic databases
HOVERGEN 311258 311246 0.01 39 Phylogenomic databases
HSSP 250661 250434 0.01 42 3D structure databases
IPI 309350 308676 0.01 40 Sequence databases
InParanoid 189538 189538 0.01 43 Phylogenomic databases
IntAct 16845 16845 <0.01 71 Protein-protein interaction databases
InterPro 63181437 22759474 2.16 1 Family and domain databases
KEGG 7902138 7711636 0.27 11 Genome annotation databases
KO 3098495 3085017 0.11 21 Phylogenomic databases
LegioList 5138 5110 <0.01 78 Organism-specific databases
Leproma 1272 1270 <0.01 88 Organism-specific databases
MEROPS 81141 81140 <0.01 53 Protein family/group databases
MGI 35077 34422 <0.01 63 Organism-specific databases
MINT 8590 8590 <0.01 74 Protein-protein interaction databases
NextBio 104044 103709 <0.01 50 Other
OMA 3889766 3889385 0.13 18 Phylogenomic databases
OrthoDB 557052 557014 0.02 34 Phylogenomic databases
PANTHER 4184001 3955873 0.14 17 Family and domain databases
PATRIC 8310247 8310154 0.28 10 Genome annotation databases
PDB 18385 10298 <0.01 68 3D structure databases
PDBsum 18152 10143 <0.01 69 3D structure databases
PHCI-2DPAGE 99 99 <0.01 98 2D gel databases
PIR 173697 140856 0.01 46 Sequence databases
PIRSF 2516170 2515493 0.09 26 Family and domain databases
PMAP-CutDB 214 214 <0.01 94 Other
PMMA-2DPAGE 2 2 <0.01 108 2D gel databases
PRIDE 476086 476086 0.02 36 Proteomic databases
PRINTS 4469756 3973785 0.15 15 Family and domain databases
PROSITE 14613027 9689583 0.50 5 Family and domain databases
Pathway_Interaction_DB 11 9 <0.01 106 Enzyme and pathway databases
PaxDb 17042 17042 <0.01 70 Proteomic databases
PeptideAtlas 144 144 <0.01 97 Proteomic databases
PeroxiBase 2558 2550 <0.01 84 Protein family/group databases
Pfam 28789057 21143842 0.98 4 Family and domain databases
PharmGKB 4279 4279 <0.01 80 Organism-specific databases
PhosphoSite 1167 1167 <0.01 89 PTM databases
PhylomeDB 151147 151147 0.01 47 Phylogenomic databases
PomBase 40 27 <0.01 102 Organism-specific databases
PptaseDB 36 34 <0.01 103 Protein family/group databases
ProDom 572760 547986 0.02 33 Family and domain databases
ProMEX 268 268 <0.01 93 Proteomic databases
ProtClustDB 2720788 2720777 0.09 24 Phylogenomic databases
ProteinModelPortal 7763747 7763747 0.27 12 3D structure databases
PseudoCAP 4539 4533 <0.01 79 Organism-specific databases
REBASE 32949 32946 <0.01 64 Protein family/group databases
REPRODUCTION-2DPAGE 84 83 <0.01 99 2D gel databases
RGD 24752 24429 <0.01 66 Organism-specific databases
Reactome 209 179 <0.01 95 Enzyme and pathway databases
RefSeq 8800149 8554497 0.30 8 Sequence databases
SGD 11 11 <0.01 105 Organism-specific databases
SMART 6511821 4935454 0.22 14 Family and domain databases
SMR 1667955 1667955 0.06 27 3D structure databases
STRING 2588334 2588334 0.09 25 Protein-protein interaction databases
SUPFAM 12032612 9897317 0.41 7 Family and domain databases
SWISS-2DPAGE 28 28 <0.01 104 2D gel databases
Siena-2DPAGE 2 2 <0.01 109 2D gel databases
TAIR 15743 15666 <0.01 72 Organism-specific databases
TCDB 2389 2377 <0.01 86 Protein family/group databases
TIGRFAMs 6651588 6065915 0.23 13 Family and domain databases
TubercuList 1991 1986 <0.01 87 Organism-specific databases
UCSC 64222 64050 <0.01 58 Genome annotation databases
UniGene 545475 514039 0.02 35 Sequence databases
UniPathway 1514470 1409900 0.05 28 Enzyme and pathway databases
VectorBase 78249 77732 <0.01 54 Genome annotation databases
World-2DPAGE 675 670 <0.01 90 2D gel databases
WormBase 42337 42219 <0.01 62 Organism-specific databases
Xenbase 25586 25469 <0.01 65 Organism-specific databases
ZFIN 45574 45448 <0.01 60 Organism-specific databases
dictyBase 7996 7774 <0.01 76 Organism-specific databases
eggNOG 2770833 2770812 0.09 23 Phylogenomic databases
euHCVdb 75267 75264 <0.01 55 Organism-specific databases
Number of explicitly cross-referenced databases: 135
5. AMINO ACID COMPOSITION
5.1 Composition in percent for the complete database
Ala (A) 8.63 Gln (Q) 3.97 Leu (L) 9.92 Ser (S) 6.65
Arg (R) 5.42 Glu (E) 6.18 Lys (K) 5.30 Thr (T) 5.56
Asn (N) 4.11 Gly (G) 7.08 Met (M) 2.47 Trp (W) 1.30
Asp (D) 5.32 His (H) 2.21 Phe (F) 4.03 Tyr (Y) 3.05
Cys (C) 1.24 Ile (I) 6.00 Pro (P) 4.67 Val (V) 6.77
Asx (B) 0.000 Glx (Z) 0 Xaa (X) 0.03
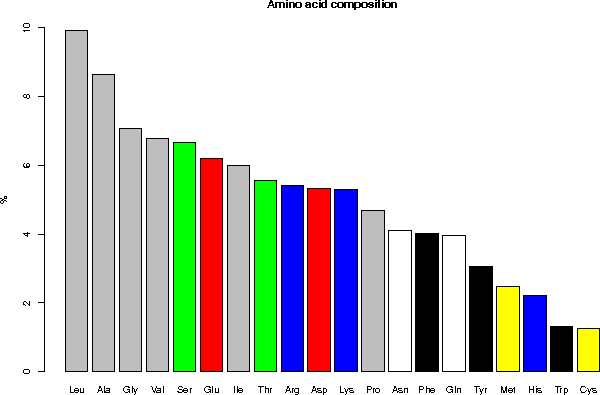 Legend: gray = aliphatic, red = acidic, green = small hydroxy,
blue = basic, black = aromatic, white = amide, yellow = sulfur
5.2 Classification of the amino acids by their frequency
Leu, Ala, Gly, Val, Ser, Glu, Ile, Thr, Arg, Asp, Lys, Pro, Asn, Phe,
Gln, Tyr, Met, His, Trp, Cys
6. MISCELLANEOUS STATISTICS
Total number of entries encoded on a Mitochondrion: 577580
Total number of entries encoded on a Plasmid: 313599
Total number of entries encoded on a Plastid: 24319
Total number of entries encoded on a Plastid; Apicoplast: 701
Total number of entries encoded on a Plastid; Chloroplast: 212091
Total number of entries encoded on a Plastid; Cyanelle: 8
Total number of entries encoded on a Plastid; Non-photosynthetic plastid: 926
Legend: gray = aliphatic, red = acidic, green = small hydroxy,
blue = basic, black = aromatic, white = amide, yellow = sulfur
5.2 Classification of the amino acids by their frequency
Leu, Ala, Gly, Val, Ser, Glu, Ile, Thr, Arg, Asp, Lys, Pro, Asn, Phe,
Gln, Tyr, Met, His, Trp, Cys
6. MISCELLANEOUS STATISTICS
Total number of entries encoded on a Mitochondrion: 577580
Total number of entries encoded on a Plasmid: 313599
Total number of entries encoded on a Plastid: 24319
Total number of entries encoded on a Plastid; Apicoplast: 701
Total number of entries encoded on a Plastid; Chloroplast: 212091
Total number of entries encoded on a Plastid; Cyanelle: 8
Total number of entries encoded on a Plastid; Non-photosynthetic plastid: 926
 2. TAXONOMIC ORIGIN
Total number of species represented in this release of UniProtKB/TrEMBL: 386951
The first twenty species represent 1753638 sequences: 6 % of the
total number of entries.
2.1 Table of the frequency of occurrence of species
Species represented 1x:16242
2x:64555
3x:34983
4x:23250
5x:14696
6x:10625
7x: 7987
8x: 6241
9x: 5054
10x: 9820
11- 20x:25669
21- 50x: 9048
51-100x: 3468
>100x: 9130
2.2 Table of the most represented species
------ --------- --------------------------------------------
Number Frequency Species
------ --------- --------------------------------------------
1 502941 Human immunodeficiency virus 1
2 180820 uncultured bacterium
3 113576 Homo sapiens (Human)
4 96951 Oryza sativa subsp. japonica (Rice)
5 78428 Hepatitis C virus
6 73724 Glycine max (Soybean) (Glycine hispida)
7 68970 Macaca mulatta (Rhesus macaque)
8 58232 Mus musculus (Mouse)
9 56116 Medicago truncatula (Barrel medic) (Medicago tribuloides)
10 54935 Hepatitis B virus (HBV)
11 54191 Danio rerio (Zebrafish) (Brachydanio rerio)
12 54089 Vitis vinifera (Grape)
13 50594 Trichomonas vaginalis
14 49230 Tetraodon nigroviridis (Spotted green pufferfish) (Chelonodon nigroviridis)
15 48878 Takifugu rubripes (Japanese pufferfish) (Fugu rubripes)
16 44540 Populus trichocarpa (Western balsam poplar)
17 43145 Callithrix jacchus (White-tufted-ear marmoset)
18 42299 Arabidopsis thaliana (Mouse-ear cress)
19 42129 Zea mays (Maize)
20 39850 Paramecium tetraurelia
21 39805 Oryza sativa subsp. indica (Rice)
22 39291 Setaria italica (Foxtail millet) (Panicum italicum)
23 38163 human gut metagenome
24 35879 Solanum lycopersicum (Tomato) (Lycopersicon esculentum)
25 35602 Ailuropoda melanoleuca (Giant panda)
26 35193 Acyrthosiphon pisum (Pea aphid)
27 35066 Caenorhabditis japonica
28 34802 Physcomitrella patens subsp. patens (Moss)
29 34453 Thalassiosira oceanica (Marine diatom)
30 34176 Drosophila melanogaster (Fruit fly)
31 33910 Rattus norvegicus (Rat)
32 33777 Sorghum bicolor (Sorghum) (Sorghum vulgare)
33 33267 Selaginella moellendorffii (Spikemoss)
34 32769 Arabidopsis lyrata subsp. lyrata (Lyre-leaved rock-cress)
35 32339 Oryza brachyantha
36 32122 Caenorhabditis remanei (Caenorhabditis vulgaris)
37 32093 Oryza glaberrima (African rice)
38 31833 Pan troglodytes (Chimpanzee)
39 31472 Sus scrofa (Pig)
40 31397 Ricinus communis (Castor bean)
41 30917 Daphnia pulex (Water flea)
42 30300 Caenorhabditis brenneri (Nematode worm)
43 30143 Brachypodium distachyon (Purple false brome) (Trachynia distachya)
44 29815 Amphimedon queenslandica (Sponge)
45 29451 Strongylocentrotus purpuratus (Purple sea urchin)
46 29315 Pristionchus pacificus
47 29178 Branchiostoma floridae (Florida lancelet) (Amphioxus)
48 29053 Oikopleura dioica (Tunicate)
49 28521 Xenopus tropicalis (Western clawed frog) (Silurana tropicalis)
50 28446 Canis familiaris (Dog) (Canis lupus familiaris)
51 28362 Escherichia coli
52 28343 Simian immunodeficiency virus (SIV)
53 28055 Gasterosteus aculeatus (Three-spined stickleback)
54 27685 Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
55 27490 Oreochromis niloticus (Nile tilapia) (Tilapia nilotica)
56 27089 Gorilla gorilla gorilla (Lowland gorilla)
57 26818 Crassostrea gigas (Pacific oyster) (Crassostrea angulata)
58 26790 Gallus gallus (Chicken)
59 25900 Oryzias latipes (Medaka fish) (Japanese ricefish)
60 25758 Loxodonta africana (African elephant)
61 25721 Phytophthora sojae (strain P6497) (Soybean stem and root rot agent)
62 25384 Bos taurus (Bovine)
63 25074 Oryctolagus cuniculus (Rabbit)
64 24879 Nematostella vectensis (Starlet sea anemone)
65 24643 Tetrahymena thermophila (strain SB210)
66 24200 Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus)
67 24059 Equus caballus (Horse)
68 23715 Ornithorhynchus anatinus (Duckbill platypus)
69 23565 Oxytricha trifallax
70 23225 Nomascus leucogenys (Northern white-cheeked gibbon) (Hylobates leucogenys)
71 23115 Perkinsus marinus (strain ATCC 50983 / TXsc)
72 22714 Monodelphis domestica (Gray short-tailed opossum)
73 22560 Sarcophilus harrisii (Tasmanian devil) (Sarcophilus laniarius)
74 22437 Caenorhabditis elegans
75 22305 Pongo abelii (Sumatran orangutan) (Pongo pygmaeus abelii)
76 22163 gut metagenome
77 21821 Latimeria chalumnae (West Indian ocean coelacanth)
78 21699 Hordeum vulgare var. distichum (Two-rowed barley)
79 21546 Heterocephalus glaber (Naked mole rat)
80 21339 Caenorhabditis briggsae
81 21086 Ixodes scapularis (Black-legged tick) (Deer tick)
82 20854 Myotis lucifugus (Little brown bat)
83 20732 Pelodiscus sinensis (Chinese softshell turtle) (Trionyx sinensis)
84 20130 Otolemur garnettii (Small-eared galago) (Garnett's greater bushbaby)
85 20114 Ciona savignyi (Pacific transparent sea squirt)
86 20069 Cavia porcellus (Guinea pig)
87 19969 Spermophilus tridecemlineatus (Thirteen-lined ground squirrel)
88 19671 Taeniopygia guttata (Zebra finch) (Poephila guttata)
89 19438 Wuchereria bancrofti
90 19320 Toxoplasma gondii
91 19255 Anolis carolinensis (Green anole) (American chameleon)
92 19200 Trypanosoma cruzi (strain CL Brener)
93 18919 Culex quinquefasciatus (Southern house mosquito) (Culex pungens)
94 18771 mine drainage metagenome
95 18706 Drosophila simulans (Fruit fly)
96 18585 Ciona intestinalis (Transparent sea squirt) (Ascidia intestinalis)
97 18121 Atta cephalotes (Leafcutter ant)
98 17839 Laccaria bicolor (strain S238N-H82 / ATCC MYA-4686) (Bicoloured deceiver)
99 17784 Fusarium oxysporum (strain Fo5176) (Panama disease fungus)
100 17599 Phytophthora infestans (strain T30-4) (Potato late blight fungus)
101 17384 Aedes aegypti (Yellowfever mosquito) (Culex aegypti)
102 17374 Bombyx mori (Silk moth)
103 17277 Nasonia vitripennis (Parasitic wasp)
104 17031 Drosophila yakuba (Fruit fly)
105 17011 Tribolium castaneum (Red flour beetle)
106 16946 Rhizopus delemar (strain RA 99-880 / ATCC MYA-4621 / FGSC 9543 / NRRL 43880)
107 16871 Meleagris gallopavo (Common turkey)
108 16714 Drosophila persimilis (Fruit fly)
109 16643 Fusarium oxysporum f. sp. lycopersici
110 16475 Drosophila pseudoobscura pseudoobscura (Fruit fly)
111 16426 Ectocarpus siliculosus (Brown alga)
112 16345 Botryotinia fuckeliana (strain T4) (Noble rot fungus) (Botrytis cinerea)
113 16306 Danaus plexippus (Monarch butterfly)
114 16263 Trichinella spiralis (Trichina worm)
115 16237 Melampsora larici-populina (strain 98AG31 / pathotype 3-4-7)
116 16188 Drosophila sechellia (Fruit fly)
117 16140 Schistosoma japonicum (Blood fluke)
118 16110 Colletotrichum higginsianum (strain IMI 349063) (Crucifer anthracnose fungus)
119 15930 Hepatitis C virus subtype 1b
120 15816 Plasmodium falciparum
121 15793 Puccinia graminis f. sp. tritici (strain CRL 75-36-700-3 / race SCCL)
122 15762 Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173)
123 15715 Naegleria gruberi (Amoeba)
124 15653 Nectria haematococca (strain 77-13-4 / ATCC MYA-4622 / FGSC 9596 / MPVI)
125 15630 Anopheles gambiae (African malaria mosquito)
126 15563 Phytophthora ramorum (Sudden oak death agent)
127 15420 Drosophila willistoni (Fruit fly)
128 15354 Loa loa (Eye worm) (Filaria loa)
129 15225 Pythium ultimum
130 15173 Hepatitis C virus subtype 1a
131 15143 Drosophila ananassae (Fruit fly)
132 15036 Harpegnathos saltator (Jerdon's jumping ant)
133 14927 Drosophila erecta (Fruit fly)
134 14851 Chlamydomonas reinhardtii (Chlamydomonas smithii)
135 14797 Camponotus floridanus (Florida carpenter ant)
136 14788 Drosophila mojavensis (Fruit fly)
137 14701 Drosophila virilis (Fruit fly)
138 14697 Plasmodium chabaudi
139 14650 Drosophila grimshawi (Fruit fly) (Idiomyia grimshawi)
140 14610 Gaeumannomyces graminis var. tritici (strain R3-111a-1)
141 14417 Volvox carteri (Green alga)
142 14339 Serpula lacrymans var. lacrymans (strain S7.3) (Dry rot fungus)
143 14336 Solenopsis invicta (Red imported fire ant) (Solenopsis wagneri)
144 14236 Sclerotinia sclerotiorum (strain ATCC 18683 / 1980 / Ss-1) (White mold)
145 13966 Acromyrmex echinatior (Panamanian leafcutter ant)
146 13867 Phanerochaete carnosa (strain HHB-10118-sp) (White-rot fungus)
147 13863 Clonorchis sinensis (Chinese liver fluke)
148 13801 Macrophomina phaseolina (strain MS6) (Charcoal rot fungus)
149 13766 Aspergillus niger (strain CBS 513.88 / FGSC A1513)
150 13648 Moniliophthora perniciosa (strain FA553 / isolate CP02)
151 13530 Trypanosoma cruzi
152 13329 Aspergillus flavus
153 13266 Coprinopsis cinerea (strain Okayama-7 / 130 / ATCC MYA-4618 / FGSC 9003)
154 13186 Mustela putorius furo (European domestic ferret) (Mustela furo)
155 13121 Schizophyllum commune (strain H4-8 / FGSC 9210) (Split gill fungus)
156 13043 Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958)
157 12983 Albugo laibachii Nc14
158 12950 Stigmatella aurantiaca (strain DW4/3-1)
159 12936 Talaromyces stipitatus (strain ATCC 10500 / CBS 375.48 / QM 6759 / NRRL 1006)
160 12900 Gibberella zeae (strain PH-1 / ATCC MYA-4620 / FGSC 9075 / NRRL 31084)
161 12722 Serpula lacrymans var. lacrymans (strain S7.9) (Dry rot fungus)
162 12696 Trypanosoma congolense (strain IL3000)
163 12682 Penicillium chrysogenum (strain ATCC 28089 / DSM 1075 / Wisconsin 54-1255)
164 12674 Schistosoma mansoni (Blood fluke)
165 12603 Xenopus laevis (African clawed frog)
166 12570 Ralstonia solanacearum (Pseudomonas solanacearum)
167 12447 Fusarium pseudograminearum (strain CS3096) (Wheat and barley crown-rot fungus)
168 12446 Leptosphaeria maculans (strain JN3 / isolate v23.1.3 / race Av1-4-5-6-7-8)
169 12440 Polysphondylium pallidum (Cellular slime mold)
170 12390 Rabies virus
171 12389 Hypocrea virens (strain Gv29-8 / FGSC 10586) (Gliocladium virens)
172 12352 Dictyostelium purpureum (Slime mold)
173 12329 uncultured archaeon
174 12152 Dictyostelium fasciculatum (strain SH3) (Slime mold)
175 11994 Pyrenophora tritici-repentis (strain Pt-1C-BFP) (Wheat tan spot fungus)
176 11993 Colletotrichum graminicola (strain M1.001 / M2 / FGSC 10212)
177 11946 Porcine reproductive and respiratory syndrome virus (PRRSV)
178 11945 Emericella nidulans
179 11927 Helicobacter pylori (Campylobacter pylori)
180 11914 Apis mellifera (Honeybee)
181 11815 Hypocrea atroviridis (strain ATCC 20476 / IMI 206040) (Trichoderma atroviride)
182 11780 Piriformospora indica (strain DSM 11827)
183 11752 Chondrocladia sp. SMF
2. TAXONOMIC ORIGIN
Total number of species represented in this release of UniProtKB/TrEMBL: 386951
The first twenty species represent 1753638 sequences: 6 % of the
total number of entries.
2.1 Table of the frequency of occurrence of species
Species represented 1x:16242
2x:64555
3x:34983
4x:23250
5x:14696
6x:10625
7x: 7987
8x: 6241
9x: 5054
10x: 9820
11- 20x:25669
21- 50x: 9048
51-100x: 3468
>100x: 9130
2.2 Table of the most represented species
------ --------- --------------------------------------------
Number Frequency Species
------ --------- --------------------------------------------
1 502941 Human immunodeficiency virus 1
2 180820 uncultured bacterium
3 113576 Homo sapiens (Human)
4 96951 Oryza sativa subsp. japonica (Rice)
5 78428 Hepatitis C virus
6 73724 Glycine max (Soybean) (Glycine hispida)
7 68970 Macaca mulatta (Rhesus macaque)
8 58232 Mus musculus (Mouse)
9 56116 Medicago truncatula (Barrel medic) (Medicago tribuloides)
10 54935 Hepatitis B virus (HBV)
11 54191 Danio rerio (Zebrafish) (Brachydanio rerio)
12 54089 Vitis vinifera (Grape)
13 50594 Trichomonas vaginalis
14 49230 Tetraodon nigroviridis (Spotted green pufferfish) (Chelonodon nigroviridis)
15 48878 Takifugu rubripes (Japanese pufferfish) (Fugu rubripes)
16 44540 Populus trichocarpa (Western balsam poplar)
17 43145 Callithrix jacchus (White-tufted-ear marmoset)
18 42299 Arabidopsis thaliana (Mouse-ear cress)
19 42129 Zea mays (Maize)
20 39850 Paramecium tetraurelia
21 39805 Oryza sativa subsp. indica (Rice)
22 39291 Setaria italica (Foxtail millet) (Panicum italicum)
23 38163 human gut metagenome
24 35879 Solanum lycopersicum (Tomato) (Lycopersicon esculentum)
25 35602 Ailuropoda melanoleuca (Giant panda)
26 35193 Acyrthosiphon pisum (Pea aphid)
27 35066 Caenorhabditis japonica
28 34802 Physcomitrella patens subsp. patens (Moss)
29 34453 Thalassiosira oceanica (Marine diatom)
30 34176 Drosophila melanogaster (Fruit fly)
31 33910 Rattus norvegicus (Rat)
32 33777 Sorghum bicolor (Sorghum) (Sorghum vulgare)
33 33267 Selaginella moellendorffii (Spikemoss)
34 32769 Arabidopsis lyrata subsp. lyrata (Lyre-leaved rock-cress)
35 32339 Oryza brachyantha
36 32122 Caenorhabditis remanei (Caenorhabditis vulgaris)
37 32093 Oryza glaberrima (African rice)
38 31833 Pan troglodytes (Chimpanzee)
39 31472 Sus scrofa (Pig)
40 31397 Ricinus communis (Castor bean)
41 30917 Daphnia pulex (Water flea)
42 30300 Caenorhabditis brenneri (Nematode worm)
43 30143 Brachypodium distachyon (Purple false brome) (Trachynia distachya)
44 29815 Amphimedon queenslandica (Sponge)
45 29451 Strongylocentrotus purpuratus (Purple sea urchin)
46 29315 Pristionchus pacificus
47 29178 Branchiostoma floridae (Florida lancelet) (Amphioxus)
48 29053 Oikopleura dioica (Tunicate)
49 28521 Xenopus tropicalis (Western clawed frog) (Silurana tropicalis)
50 28446 Canis familiaris (Dog) (Canis lupus familiaris)
51 28362 Escherichia coli
52 28343 Simian immunodeficiency virus (SIV)
53 28055 Gasterosteus aculeatus (Three-spined stickleback)
54 27685 Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
55 27490 Oreochromis niloticus (Nile tilapia) (Tilapia nilotica)
56 27089 Gorilla gorilla gorilla (Lowland gorilla)
57 26818 Crassostrea gigas (Pacific oyster) (Crassostrea angulata)
58 26790 Gallus gallus (Chicken)
59 25900 Oryzias latipes (Medaka fish) (Japanese ricefish)
60 25758 Loxodonta africana (African elephant)
61 25721 Phytophthora sojae (strain P6497) (Soybean stem and root rot agent)
62 25384 Bos taurus (Bovine)
63 25074 Oryctolagus cuniculus (Rabbit)
64 24879 Nematostella vectensis (Starlet sea anemone)
65 24643 Tetrahymena thermophila (strain SB210)
66 24200 Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus)
67 24059 Equus caballus (Horse)
68 23715 Ornithorhynchus anatinus (Duckbill platypus)
69 23565 Oxytricha trifallax
70 23225 Nomascus leucogenys (Northern white-cheeked gibbon) (Hylobates leucogenys)
71 23115 Perkinsus marinus (strain ATCC 50983 / TXsc)
72 22714 Monodelphis domestica (Gray short-tailed opossum)
73 22560 Sarcophilus harrisii (Tasmanian devil) (Sarcophilus laniarius)
74 22437 Caenorhabditis elegans
75 22305 Pongo abelii (Sumatran orangutan) (Pongo pygmaeus abelii)
76 22163 gut metagenome
77 21821 Latimeria chalumnae (West Indian ocean coelacanth)
78 21699 Hordeum vulgare var. distichum (Two-rowed barley)
79 21546 Heterocephalus glaber (Naked mole rat)
80 21339 Caenorhabditis briggsae
81 21086 Ixodes scapularis (Black-legged tick) (Deer tick)
82 20854 Myotis lucifugus (Little brown bat)
83 20732 Pelodiscus sinensis (Chinese softshell turtle) (Trionyx sinensis)
84 20130 Otolemur garnettii (Small-eared galago) (Garnett's greater bushbaby)
85 20114 Ciona savignyi (Pacific transparent sea squirt)
86 20069 Cavia porcellus (Guinea pig)
87 19969 Spermophilus tridecemlineatus (Thirteen-lined ground squirrel)
88 19671 Taeniopygia guttata (Zebra finch) (Poephila guttata)
89 19438 Wuchereria bancrofti
90 19320 Toxoplasma gondii
91 19255 Anolis carolinensis (Green anole) (American chameleon)
92 19200 Trypanosoma cruzi (strain CL Brener)
93 18919 Culex quinquefasciatus (Southern house mosquito) (Culex pungens)
94 18771 mine drainage metagenome
95 18706 Drosophila simulans (Fruit fly)
96 18585 Ciona intestinalis (Transparent sea squirt) (Ascidia intestinalis)
97 18121 Atta cephalotes (Leafcutter ant)
98 17839 Laccaria bicolor (strain S238N-H82 / ATCC MYA-4686) (Bicoloured deceiver)
99 17784 Fusarium oxysporum (strain Fo5176) (Panama disease fungus)
100 17599 Phytophthora infestans (strain T30-4) (Potato late blight fungus)
101 17384 Aedes aegypti (Yellowfever mosquito) (Culex aegypti)
102 17374 Bombyx mori (Silk moth)
103 17277 Nasonia vitripennis (Parasitic wasp)
104 17031 Drosophila yakuba (Fruit fly)
105 17011 Tribolium castaneum (Red flour beetle)
106 16946 Rhizopus delemar (strain RA 99-880 / ATCC MYA-4621 / FGSC 9543 / NRRL 43880)
107 16871 Meleagris gallopavo (Common turkey)
108 16714 Drosophila persimilis (Fruit fly)
109 16643 Fusarium oxysporum f. sp. lycopersici
110 16475 Drosophila pseudoobscura pseudoobscura (Fruit fly)
111 16426 Ectocarpus siliculosus (Brown alga)
112 16345 Botryotinia fuckeliana (strain T4) (Noble rot fungus) (Botrytis cinerea)
113 16306 Danaus plexippus (Monarch butterfly)
114 16263 Trichinella spiralis (Trichina worm)
115 16237 Melampsora larici-populina (strain 98AG31 / pathotype 3-4-7)
116 16188 Drosophila sechellia (Fruit fly)
117 16140 Schistosoma japonicum (Blood fluke)
118 16110 Colletotrichum higginsianum (strain IMI 349063) (Crucifer anthracnose fungus)
119 15930 Hepatitis C virus subtype 1b
120 15816 Plasmodium falciparum
121 15793 Puccinia graminis f. sp. tritici (strain CRL 75-36-700-3 / race SCCL)
122 15762 Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173)
123 15715 Naegleria gruberi (Amoeba)
124 15653 Nectria haematococca (strain 77-13-4 / ATCC MYA-4622 / FGSC 9596 / MPVI)
125 15630 Anopheles gambiae (African malaria mosquito)
126 15563 Phytophthora ramorum (Sudden oak death agent)
127 15420 Drosophila willistoni (Fruit fly)
128 15354 Loa loa (Eye worm) (Filaria loa)
129 15225 Pythium ultimum
130 15173 Hepatitis C virus subtype 1a
131 15143 Drosophila ananassae (Fruit fly)
132 15036 Harpegnathos saltator (Jerdon's jumping ant)
133 14927 Drosophila erecta (Fruit fly)
134 14851 Chlamydomonas reinhardtii (Chlamydomonas smithii)
135 14797 Camponotus floridanus (Florida carpenter ant)
136 14788 Drosophila mojavensis (Fruit fly)
137 14701 Drosophila virilis (Fruit fly)
138 14697 Plasmodium chabaudi
139 14650 Drosophila grimshawi (Fruit fly) (Idiomyia grimshawi)
140 14610 Gaeumannomyces graminis var. tritici (strain R3-111a-1)
141 14417 Volvox carteri (Green alga)
142 14339 Serpula lacrymans var. lacrymans (strain S7.3) (Dry rot fungus)
143 14336 Solenopsis invicta (Red imported fire ant) (Solenopsis wagneri)
144 14236 Sclerotinia sclerotiorum (strain ATCC 18683 / 1980 / Ss-1) (White mold)
145 13966 Acromyrmex echinatior (Panamanian leafcutter ant)
146 13867 Phanerochaete carnosa (strain HHB-10118-sp) (White-rot fungus)
147 13863 Clonorchis sinensis (Chinese liver fluke)
148 13801 Macrophomina phaseolina (strain MS6) (Charcoal rot fungus)
149 13766 Aspergillus niger (strain CBS 513.88 / FGSC A1513)
150 13648 Moniliophthora perniciosa (strain FA553 / isolate CP02)
151 13530 Trypanosoma cruzi
152 13329 Aspergillus flavus
153 13266 Coprinopsis cinerea (strain Okayama-7 / 130 / ATCC MYA-4618 / FGSC 9003)
154 13186 Mustela putorius furo (European domestic ferret) (Mustela furo)
155 13121 Schizophyllum commune (strain H4-8 / FGSC 9210) (Split gill fungus)
156 13043 Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958)
157 12983 Albugo laibachii Nc14
158 12950 Stigmatella aurantiaca (strain DW4/3-1)
159 12936 Talaromyces stipitatus (strain ATCC 10500 / CBS 375.48 / QM 6759 / NRRL 1006)
160 12900 Gibberella zeae (strain PH-1 / ATCC MYA-4620 / FGSC 9075 / NRRL 31084)
161 12722 Serpula lacrymans var. lacrymans (strain S7.9) (Dry rot fungus)
162 12696 Trypanosoma congolense (strain IL3000)
163 12682 Penicillium chrysogenum (strain ATCC 28089 / DSM 1075 / Wisconsin 54-1255)
164 12674 Schistosoma mansoni (Blood fluke)
165 12603 Xenopus laevis (African clawed frog)
166 12570 Ralstonia solanacearum (Pseudomonas solanacearum)
167 12447 Fusarium pseudograminearum (strain CS3096) (Wheat and barley crown-rot fungus)
168 12446 Leptosphaeria maculans (strain JN3 / isolate v23.1.3 / race Av1-4-5-6-7-8)
169 12440 Polysphondylium pallidum (Cellular slime mold)
170 12390 Rabies virus
171 12389 Hypocrea virens (strain Gv29-8 / FGSC 10586) (Gliocladium virens)
172 12352 Dictyostelium purpureum (Slime mold)
173 12329 uncultured archaeon
174 12152 Dictyostelium fasciculatum (strain SH3) (Slime mold)
175 11994 Pyrenophora tritici-repentis (strain Pt-1C-BFP) (Wheat tan spot fungus)
176 11993 Colletotrichum graminicola (strain M1.001 / M2 / FGSC 10212)
177 11946 Porcine reproductive and respiratory syndrome virus (PRRSV)
178 11945 Emericella nidulans
179 11927 Helicobacter pylori (Campylobacter pylori)
180 11914 Apis mellifera (Honeybee)
181 11815 Hypocrea atroviridis (strain ATCC 20476 / IMI 206040) (Trichoderma atroviride)
182 11780 Piriformospora indica (strain DSM 11827)
183 11752 Chondrocladia sp. SMF Kingdom sequences (% of the database)
Archaea 408842 ( 1%)
Bacteria 20361597 ( 70%)
Eukaryota 6846383 ( 23%)
Viruses 1547692 ( 5%)
Other 102424 ( <1%)
Within Eukaryota:
Kingdom sequences (% of the database)
Archaea 408842 ( 1%)
Bacteria 20361597 ( 70%)
Eukaryota 6846383 ( 23%)
Viruses 1547692 ( 5%)
Other 102424 ( <1%)
Within Eukaryota:
 Category sequences (% of Eukaryota) (% of the complete database)
Human 113612 ( 2%) ( 0%)
Other Mammalia 845085 ( 12%) ( 3%)
Other Vertebrata 753401 ( 11%) ( 3%)
Viridiplantae 1317778 ( 19%) ( 5%)
Fungi 1510587 ( 22%) ( 5%)
Insecta 782338 ( 11%) ( 3%)
Nematoda 252562 ( 4%) ( 1%)
Other 1271020 ( 19%) ( 4%)
3. SEQUENCE SIZE
Repartition of the sequences by size (excluding fragments)
From To Number From To Number
1- 50 755730 1001-1100 168936
51- 100 2504899 1101-1200 118418
101- 150 2798075 1201-1300 83249
151- 200 2715002 1301-1400 53034
201- 250 2729580 1401-1500 42871
251- 300 2645019 1501-1600 29671
301- 350 2401964 1601-1700 22537
351- 400 1820598 1701-1800 17087
401- 450 1569876 1801-1900 14194
451- 500 1287032 1901-2000 12118
501- 550 851336 2001-2100 9497
551- 600 655543 2101-2200 9743
601- 650 479546 2201-2300 7592
651- 700 376441 2301-2400 6087
701- 750 317101 2401-2500 5197
751- 800 279539 >2500 42031
801- 850 213365
851- 900 190510
901- 950 132149
951-1000 97011
Category sequences (% of Eukaryota) (% of the complete database)
Human 113612 ( 2%) ( 0%)
Other Mammalia 845085 ( 12%) ( 3%)
Other Vertebrata 753401 ( 11%) ( 3%)
Viridiplantae 1317778 ( 19%) ( 5%)
Fungi 1510587 ( 22%) ( 5%)
Insecta 782338 ( 11%) ( 3%)
Nematoda 252562 ( 4%) ( 1%)
Other 1271020 ( 19%) ( 4%)
3. SEQUENCE SIZE
Repartition of the sequences by size (excluding fragments)
From To Number From To Number
1- 50 755730 1001-1100 168936
51- 100 2504899 1101-1200 118418
101- 150 2798075 1201-1300 83249
151- 200 2715002 1301-1400 53034
201- 250 2729580 1401-1500 42871
251- 300 2645019 1501-1600 29671
301- 350 2401964 1601-1700 22537
351- 400 1820598 1701-1800 17087
401- 450 1569876 1801-1900 14194
451- 500 1287032 1901-2000 12118
501- 550 851336 2001-2100 9497
551- 600 655543 2101-2200 9743
601- 650 479546 2201-2300 7592
651- 700 376441 2301-2400 6087
701- 750 317101 2401-2500 5197
751- 800 279539 >2500 42031
801- 850 213365
851- 900 190510
901- 950 132149
951-1000 97011
 The average sequence length in UniProtKB/TrEMBL is 322 amino acids.
The shortest sequence is G0XMK1_9MYRT: 1 amino acids.
The longest sequence is Q3ASY8_CHLCH: 36805 amino acids.
4. STATISTICS FOR SOME LINE TYPES
The following table summarizes the total number of some UniProtKB/TrEMBL lines,
as well as the number of entries with at least one such line, and the
frequency of the lines.
Total Number of Average
Line type / subtype number entries per entry
--------------------------------- -------- --------- ---------
References (RL) 35659949 1.22
Submitted to EMBL/GenBank/DDBJ 19645186 18124632 0.67
Journal 14507716 13660992 0.50
Submitted to other databases 1490660 1481406 0.05
Thesis 9861 9803 <0.01
Book citation 6506 6457 <0.01
Unpublished observations 19 19 <0.01
Patent 1 1 <0.01
Total number of distinct authors cited in UniProtKB/TrEMBL: 456562
Total Number of Average
Line type / subtype number entries per entry Rank
--------------------------------- -------- --------- --------- ----
Comments (CC) 36180072 1.24
CATALYTIC ACTIVITY 3133293 2851026 0.11 4
CAUTION 13393492 13393247 0.46 1
COFACTOR 1186735 1098197 0.04 8
DOMAIN 118504 113749 <0.01 9
FUNCTION 3425325 3205049 0.12 3
INTERACTION 690 690 <0.01 11
MISCELLANEOUS 82701 82605 <0.01 10
PATHWAY 1552547 1411355 0.05 7
SIMILARITY 8834143 7669541 0.30 2
SUBCELLULAR LOCATION 2759375 2633226 0.09 5
SUBUNIT 1693267 1673876 0.06 6
Total number of comment topics: 11
Total Number of Average
Line type / subtype number entries per entry Rank
--------------------------------- -------- --------- --------- ----
Features (FT) 7381377 0.25
CHAIN 774905 642346 0.03 2
NON_TER 5990131 3805013 0.20 1
SIGNAL 615484 612214 0.02 3
TRANSIT 857 856 <0.01 4
Total number of feature keys: 4
Total Number of Average
Line type / subtype number entries per entry Rank Category
--------------------------------- -------- --------- --------- ---- -------------------------------------------
Cross-references (DR) 330404216 11.29
AGD 2525 2525 <0.01 85 Organism-specific databases
ANU-2DPAGE 52 52 <0.01 101 2D gel databases
Allergome 3024 2409 <0.01 81 Protein family/group databases
ArachnoServer 66 66 <0.01 100 Organism-specific databases
ArrayExpress 86858 86858 <0.01 52 Gene expression databases
BRENDA 2680 2651 <0.01 83 Enzyme and pathway databases
Bgee 118824 118824 <0.01 49 Gene expression databases
BioCyc 3585778 3547066 0.12 20 Enzyme and pathway databases
CAZy 74131 69652 <0.01 56 Protein family/group databases
CGD 7064 7064 <0.01 77 Organism-specific databases
COMPLUYEAST-2DPAGE 5 5 <0.01 107 2D gel databases
CTD 322062 320469 0.01 38 Organism-specific databases
ChEMBL 576 576 <0.01 92 Other
ConoServer 160 160 <0.01 96 Organism-specific databases
DIP 2782 2777 <0.01 82 Protein-protein interaction databases
DNASU 43665 43327 <0.01 61 Protocols and materials databases
EMBL 32113816 28423223 1.10 3 Sequence databases
Ensembl 959177 943419 0.03 29 Genome annotation databases
EnsemblBacteria 834798 800722 0.03 30 Genome annotation databases
EnsemblFungi 262813 261328 0.01 41 Genome annotation databases
EnsemblMetazoa 629747 614533 0.02 32 Genome annotation databases
EnsemblPlants 425229 405703 0.01 37 Genome annotation databases
EnsemblProtists 126697 125197 <0.01 48 Genome annotation databases
EuPathDB 178957 178954 0.01 45 Organism-specific databases
EvolutionaryTrace 8177 8177 <0.01 75 Other
FlyBase 182089 180690 0.01 44 Organism-specific databases
GO 59000915 18239516 2.02 2 Ontologies
Gene3D 12548459 9987923 0.43 6 Family and domain databases
GeneID 8760277 8545494 0.30 9 Genome annotation databases
GeneTree 798347 798164 0.03 31 Phylogenomic databases
Genevestigator 93341 93333 <0.01 51 Gene expression databases
GenoList 14735 14462 <0.01 73 Organism-specific databases
GenomeRNAi 21434 21434 <0.01 67 Other
GenomeReviews 4251570 4152775 0.15 16 Genome annotation databases
Gramene 67608 67608 <0.01 57 Organism-specific databases
H-InvDB 626 478 <0.01 91 Organism-specific databases
HAMAP 2916833 2880798 0.10 22 Family and domain databases
HGNC 48958 48878 <0.01 59 Organism-specific databases
HOGENOM 3658839 3658792 0.13 19 Phylogenomic databases
HOVERGEN 311258 311246 0.01 39 Phylogenomic databases
HSSP 250661 250434 0.01 42 3D structure databases
IPI 309350 308676 0.01 40 Sequence databases
InParanoid 189538 189538 0.01 43 Phylogenomic databases
IntAct 16845 16845 <0.01 71 Protein-protein interaction databases
InterPro 63181437 22759474 2.16 1 Family and domain databases
KEGG 7902138 7711636 0.27 11 Genome annotation databases
KO 3098495 3085017 0.11 21 Phylogenomic databases
LegioList 5138 5110 <0.01 78 Organism-specific databases
Leproma 1272 1270 <0.01 88 Organism-specific databases
MEROPS 81141 81140 <0.01 53 Protein family/group databases
MGI 35077 34422 <0.01 63 Organism-specific databases
MINT 8590 8590 <0.01 74 Protein-protein interaction databases
NextBio 104044 103709 <0.01 50 Other
OMA 3889766 3889385 0.13 18 Phylogenomic databases
OrthoDB 557052 557014 0.02 34 Phylogenomic databases
PANTHER 4184001 3955873 0.14 17 Family and domain databases
PATRIC 8310247 8310154 0.28 10 Genome annotation databases
PDB 18385 10298 <0.01 68 3D structure databases
PDBsum 18152 10143 <0.01 69 3D structure databases
PHCI-2DPAGE 99 99 <0.01 98 2D gel databases
PIR 173697 140856 0.01 46 Sequence databases
PIRSF 2516170 2515493 0.09 26 Family and domain databases
PMAP-CutDB 214 214 <0.01 94 Other
PMMA-2DPAGE 2 2 <0.01 108 2D gel databases
PRIDE 476086 476086 0.02 36 Proteomic databases
PRINTS 4469756 3973785 0.15 15 Family and domain databases
PROSITE 14613027 9689583 0.50 5 Family and domain databases
Pathway_Interaction_DB 11 9 <0.01 106 Enzyme and pathway databases
PaxDb 17042 17042 <0.01 70 Proteomic databases
PeptideAtlas 144 144 <0.01 97 Proteomic databases
PeroxiBase 2558 2550 <0.01 84 Protein family/group databases
Pfam 28789057 21143842 0.98 4 Family and domain databases
PharmGKB 4279 4279 <0.01 80 Organism-specific databases
PhosphoSite 1167 1167 <0.01 89 PTM databases
PhylomeDB 151147 151147 0.01 47 Phylogenomic databases
PomBase 40 27 <0.01 102 Organism-specific databases
PptaseDB 36 34 <0.01 103 Protein family/group databases
ProDom 572760 547986 0.02 33 Family and domain databases
ProMEX 268 268 <0.01 93 Proteomic databases
ProtClustDB 2720788 2720777 0.09 24 Phylogenomic databases
ProteinModelPortal 7763747 7763747 0.27 12 3D structure databases
PseudoCAP 4539 4533 <0.01 79 Organism-specific databases
REBASE 32949 32946 <0.01 64 Protein family/group databases
REPRODUCTION-2DPAGE 84 83 <0.01 99 2D gel databases
RGD 24752 24429 <0.01 66 Organism-specific databases
Reactome 209 179 <0.01 95 Enzyme and pathway databases
RefSeq 8800149 8554497 0.30 8 Sequence databases
SGD 11 11 <0.01 105 Organism-specific databases
SMART 6511821 4935454 0.22 14 Family and domain databases
SMR 1667955 1667955 0.06 27 3D structure databases
STRING 2588334 2588334 0.09 25 Protein-protein interaction databases
SUPFAM 12032612 9897317 0.41 7 Family and domain databases
SWISS-2DPAGE 28 28 <0.01 104 2D gel databases
Siena-2DPAGE 2 2 <0.01 109 2D gel databases
TAIR 15743 15666 <0.01 72 Organism-specific databases
TCDB 2389 2377 <0.01 86 Protein family/group databases
TIGRFAMs 6651588 6065915 0.23 13 Family and domain databases
TubercuList 1991 1986 <0.01 87 Organism-specific databases
UCSC 64222 64050 <0.01 58 Genome annotation databases
UniGene 545475 514039 0.02 35 Sequence databases
UniPathway 1514470 1409900 0.05 28 Enzyme and pathway databases
VectorBase 78249 77732 <0.01 54 Genome annotation databases
World-2DPAGE 675 670 <0.01 90 2D gel databases
WormBase 42337 42219 <0.01 62 Organism-specific databases
Xenbase 25586 25469 <0.01 65 Organism-specific databases
ZFIN 45574 45448 <0.01 60 Organism-specific databases
dictyBase 7996 7774 <0.01 76 Organism-specific databases
eggNOG 2770833 2770812 0.09 23 Phylogenomic databases
euHCVdb 75267 75264 <0.01 55 Organism-specific databases
Number of explicitly cross-referenced databases: 135
5. AMINO ACID COMPOSITION
5.1 Composition in percent for the complete database
Ala (A) 8.63 Gln (Q) 3.97 Leu (L) 9.92 Ser (S) 6.65
Arg (R) 5.42 Glu (E) 6.18 Lys (K) 5.30 Thr (T) 5.56
Asn (N) 4.11 Gly (G) 7.08 Met (M) 2.47 Trp (W) 1.30
Asp (D) 5.32 His (H) 2.21 Phe (F) 4.03 Tyr (Y) 3.05
Cys (C) 1.24 Ile (I) 6.00 Pro (P) 4.67 Val (V) 6.77
Asx (B) 0.000 Glx (Z) 0 Xaa (X) 0.03
The average sequence length in UniProtKB/TrEMBL is 322 amino acids.
The shortest sequence is G0XMK1_9MYRT: 1 amino acids.
The longest sequence is Q3ASY8_CHLCH: 36805 amino acids.
4. STATISTICS FOR SOME LINE TYPES
The following table summarizes the total number of some UniProtKB/TrEMBL lines,
as well as the number of entries with at least one such line, and the
frequency of the lines.
Total Number of Average
Line type / subtype number entries per entry
--------------------------------- -------- --------- ---------
References (RL) 35659949 1.22
Submitted to EMBL/GenBank/DDBJ 19645186 18124632 0.67
Journal 14507716 13660992 0.50
Submitted to other databases 1490660 1481406 0.05
Thesis 9861 9803 <0.01
Book citation 6506 6457 <0.01
Unpublished observations 19 19 <0.01
Patent 1 1 <0.01
Total number of distinct authors cited in UniProtKB/TrEMBL: 456562
Total Number of Average
Line type / subtype number entries per entry Rank
--------------------------------- -------- --------- --------- ----
Comments (CC) 36180072 1.24
CATALYTIC ACTIVITY 3133293 2851026 0.11 4
CAUTION 13393492 13393247 0.46 1
COFACTOR 1186735 1098197 0.04 8
DOMAIN 118504 113749 <0.01 9
FUNCTION 3425325 3205049 0.12 3
INTERACTION 690 690 <0.01 11
MISCELLANEOUS 82701 82605 <0.01 10
PATHWAY 1552547 1411355 0.05 7
SIMILARITY 8834143 7669541 0.30 2
SUBCELLULAR LOCATION 2759375 2633226 0.09 5
SUBUNIT 1693267 1673876 0.06 6
Total number of comment topics: 11
Total Number of Average
Line type / subtype number entries per entry Rank
--------------------------------- -------- --------- --------- ----
Features (FT) 7381377 0.25
CHAIN 774905 642346 0.03 2
NON_TER 5990131 3805013 0.20 1
SIGNAL 615484 612214 0.02 3
TRANSIT 857 856 <0.01 4
Total number of feature keys: 4
Total Number of Average
Line type / subtype number entries per entry Rank Category
--------------------------------- -------- --------- --------- ---- -------------------------------------------
Cross-references (DR) 330404216 11.29
AGD 2525 2525 <0.01 85 Organism-specific databases
ANU-2DPAGE 52 52 <0.01 101 2D gel databases
Allergome 3024 2409 <0.01 81 Protein family/group databases
ArachnoServer 66 66 <0.01 100 Organism-specific databases
ArrayExpress 86858 86858 <0.01 52 Gene expression databases
BRENDA 2680 2651 <0.01 83 Enzyme and pathway databases
Bgee 118824 118824 <0.01 49 Gene expression databases
BioCyc 3585778 3547066 0.12 20 Enzyme and pathway databases
CAZy 74131 69652 <0.01 56 Protein family/group databases
CGD 7064 7064 <0.01 77 Organism-specific databases
COMPLUYEAST-2DPAGE 5 5 <0.01 107 2D gel databases
CTD 322062 320469 0.01 38 Organism-specific databases
ChEMBL 576 576 <0.01 92 Other
ConoServer 160 160 <0.01 96 Organism-specific databases
DIP 2782 2777 <0.01 82 Protein-protein interaction databases
DNASU 43665 43327 <0.01 61 Protocols and materials databases
EMBL 32113816 28423223 1.10 3 Sequence databases
Ensembl 959177 943419 0.03 29 Genome annotation databases
EnsemblBacteria 834798 800722 0.03 30 Genome annotation databases
EnsemblFungi 262813 261328 0.01 41 Genome annotation databases
EnsemblMetazoa 629747 614533 0.02 32 Genome annotation databases
EnsemblPlants 425229 405703 0.01 37 Genome annotation databases
EnsemblProtists 126697 125197 <0.01 48 Genome annotation databases
EuPathDB 178957 178954 0.01 45 Organism-specific databases
EvolutionaryTrace 8177 8177 <0.01 75 Other
FlyBase 182089 180690 0.01 44 Organism-specific databases
GO 59000915 18239516 2.02 2 Ontologies
Gene3D 12548459 9987923 0.43 6 Family and domain databases
GeneID 8760277 8545494 0.30 9 Genome annotation databases
GeneTree 798347 798164 0.03 31 Phylogenomic databases
Genevestigator 93341 93333 <0.01 51 Gene expression databases
GenoList 14735 14462 <0.01 73 Organism-specific databases
GenomeRNAi 21434 21434 <0.01 67 Other
GenomeReviews 4251570 4152775 0.15 16 Genome annotation databases
Gramene 67608 67608 <0.01 57 Organism-specific databases
H-InvDB 626 478 <0.01 91 Organism-specific databases
HAMAP 2916833 2880798 0.10 22 Family and domain databases
HGNC 48958 48878 <0.01 59 Organism-specific databases
HOGENOM 3658839 3658792 0.13 19 Phylogenomic databases
HOVERGEN 311258 311246 0.01 39 Phylogenomic databases
HSSP 250661 250434 0.01 42 3D structure databases
IPI 309350 308676 0.01 40 Sequence databases
InParanoid 189538 189538 0.01 43 Phylogenomic databases
IntAct 16845 16845 <0.01 71 Protein-protein interaction databases
InterPro 63181437 22759474 2.16 1 Family and domain databases
KEGG 7902138 7711636 0.27 11 Genome annotation databases
KO 3098495 3085017 0.11 21 Phylogenomic databases
LegioList 5138 5110 <0.01 78 Organism-specific databases
Leproma 1272 1270 <0.01 88 Organism-specific databases
MEROPS 81141 81140 <0.01 53 Protein family/group databases
MGI 35077 34422 <0.01 63 Organism-specific databases
MINT 8590 8590 <0.01 74 Protein-protein interaction databases
NextBio 104044 103709 <0.01 50 Other
OMA 3889766 3889385 0.13 18 Phylogenomic databases
OrthoDB 557052 557014 0.02 34 Phylogenomic databases
PANTHER 4184001 3955873 0.14 17 Family and domain databases
PATRIC 8310247 8310154 0.28 10 Genome annotation databases
PDB 18385 10298 <0.01 68 3D structure databases
PDBsum 18152 10143 <0.01 69 3D structure databases
PHCI-2DPAGE 99 99 <0.01 98 2D gel databases
PIR 173697 140856 0.01 46 Sequence databases
PIRSF 2516170 2515493 0.09 26 Family and domain databases
PMAP-CutDB 214 214 <0.01 94 Other
PMMA-2DPAGE 2 2 <0.01 108 2D gel databases
PRIDE 476086 476086 0.02 36 Proteomic databases
PRINTS 4469756 3973785 0.15 15 Family and domain databases
PROSITE 14613027 9689583 0.50 5 Family and domain databases
Pathway_Interaction_DB 11 9 <0.01 106 Enzyme and pathway databases
PaxDb 17042 17042 <0.01 70 Proteomic databases
PeptideAtlas 144 144 <0.01 97 Proteomic databases
PeroxiBase 2558 2550 <0.01 84 Protein family/group databases
Pfam 28789057 21143842 0.98 4 Family and domain databases
PharmGKB 4279 4279 <0.01 80 Organism-specific databases
PhosphoSite 1167 1167 <0.01 89 PTM databases
PhylomeDB 151147 151147 0.01 47 Phylogenomic databases
PomBase 40 27 <0.01 102 Organism-specific databases
PptaseDB 36 34 <0.01 103 Protein family/group databases
ProDom 572760 547986 0.02 33 Family and domain databases
ProMEX 268 268 <0.01 93 Proteomic databases
ProtClustDB 2720788 2720777 0.09 24 Phylogenomic databases
ProteinModelPortal 7763747 7763747 0.27 12 3D structure databases
PseudoCAP 4539 4533 <0.01 79 Organism-specific databases
REBASE 32949 32946 <0.01 64 Protein family/group databases
REPRODUCTION-2DPAGE 84 83 <0.01 99 2D gel databases
RGD 24752 24429 <0.01 66 Organism-specific databases
Reactome 209 179 <0.01 95 Enzyme and pathway databases
RefSeq 8800149 8554497 0.30 8 Sequence databases
SGD 11 11 <0.01 105 Organism-specific databases
SMART 6511821 4935454 0.22 14 Family and domain databases
SMR 1667955 1667955 0.06 27 3D structure databases
STRING 2588334 2588334 0.09 25 Protein-protein interaction databases
SUPFAM 12032612 9897317 0.41 7 Family and domain databases
SWISS-2DPAGE 28 28 <0.01 104 2D gel databases
Siena-2DPAGE 2 2 <0.01 109 2D gel databases
TAIR 15743 15666 <0.01 72 Organism-specific databases
TCDB 2389 2377 <0.01 86 Protein family/group databases
TIGRFAMs 6651588 6065915 0.23 13 Family and domain databases
TubercuList 1991 1986 <0.01 87 Organism-specific databases
UCSC 64222 64050 <0.01 58 Genome annotation databases
UniGene 545475 514039 0.02 35 Sequence databases
UniPathway 1514470 1409900 0.05 28 Enzyme and pathway databases
VectorBase 78249 77732 <0.01 54 Genome annotation databases
World-2DPAGE 675 670 <0.01 90 2D gel databases
WormBase 42337 42219 <0.01 62 Organism-specific databases
Xenbase 25586 25469 <0.01 65 Organism-specific databases
ZFIN 45574 45448 <0.01 60 Organism-specific databases
dictyBase 7996 7774 <0.01 76 Organism-specific databases
eggNOG 2770833 2770812 0.09 23 Phylogenomic databases
euHCVdb 75267 75264 <0.01 55 Organism-specific databases
Number of explicitly cross-referenced databases: 135
5. AMINO ACID COMPOSITION
5.1 Composition in percent for the complete database
Ala (A) 8.63 Gln (Q) 3.97 Leu (L) 9.92 Ser (S) 6.65
Arg (R) 5.42 Glu (E) 6.18 Lys (K) 5.30 Thr (T) 5.56
Asn (N) 4.11 Gly (G) 7.08 Met (M) 2.47 Trp (W) 1.30
Asp (D) 5.32 His (H) 2.21 Phe (F) 4.03 Tyr (Y) 3.05
Cys (C) 1.24 Ile (I) 6.00 Pro (P) 4.67 Val (V) 6.77
Asx (B) 0.000 Glx (Z) 0 Xaa (X) 0.03
 Legend: gray = aliphatic, red = acidic, green = small hydroxy,
blue = basic, black = aromatic, white = amide, yellow = sulfur
5.2 Classification of the amino acids by their frequency
Leu, Ala, Gly, Val, Ser, Glu, Ile, Thr, Arg, Asp, Lys, Pro, Asn, Phe,
Gln, Tyr, Met, His, Trp, Cys
6. MISCELLANEOUS STATISTICS
Total number of entries encoded on a Mitochondrion: 577580
Total number of entries encoded on a Plasmid: 313599
Total number of entries encoded on a Plastid: 24319
Total number of entries encoded on a Plastid; Apicoplast: 701
Total number of entries encoded on a Plastid; Chloroplast: 212091
Total number of entries encoded on a Plastid; Cyanelle: 8
Total number of entries encoded on a Plastid; Non-photosynthetic plastid: 926
Legend: gray = aliphatic, red = acidic, green = small hydroxy,
blue = basic, black = aromatic, white = amide, yellow = sulfur
5.2 Classification of the amino acids by their frequency
Leu, Ala, Gly, Val, Ser, Glu, Ile, Thr, Arg, Asp, Lys, Pro, Asn, Phe,
Gln, Tyr, Met, His, Trp, Cys
6. MISCELLANEOUS STATISTICS
Total number of entries encoded on a Mitochondrion: 577580
Total number of entries encoded on a Plasmid: 313599
Total number of entries encoded on a Plastid: 24319
Total number of entries encoded on a Plastid; Apicoplast: 701
Total number of entries encoded on a Plastid; Chloroplast: 212091
Total number of entries encoded on a Plastid; Cyanelle: 8
Total number of entries encoded on a Plastid; Non-photosynthetic plastid: 926